Abstract
Test environment evaluation has become an increasingly important issue in plant breeding. In the context of indirect selection, a test environment can be characterized by two parameters: the heritability in the test environment and its genetic correlation with the target environment. In the context of GGE biplot analysis, a test environment is similarly characterized by two parameters: its discrimination power and its similarity with other environments. This paper investigates the relationships between GGE biplots based on different data scaling methods and the theory of indirect selection, and introduces a heritability-adjusted (HA) GGE biplot. We demonstrate that the vector length of an environment in the HA-GGE biplot approximates the square root heritability (\( \sqrt H \)) within the environment and that the cosine of the angle between the vectors of two environments approximates the genetic correlation (r) between them. Moreover, projections of vectors of test environments onto that of a target environment approximate values of \( r\sqrt H \), which are proportional to the predicted genetic gain expected in the target environment from indirect selection in the test environments at a constant selection intensity. Thus, the HA-GGE biplot graphically displays the relative utility of environments in terms of selection response. Therefore, the HA-GGE biplot is the preferred GGE biplot for test environment evaluation. It is also the appropriate GGE biplot for genotype evaluation because it weights information from the different environments proportional to their within-environment square root heritability. Approximation of the HA-GGE biplot by other types of GGE biplots was discussed.
Similar content being viewed by others
Test environment evaluation is an important research area in plant breeding, because appropriate choice of test environments can reduce test cost and improve breeding efficiency. Test environment evaluation has been discussed mainly in the context of indirect selection (Cooper et al. 1996; Guillen-Portal et al. 2004). According to Falconer and Mackay (1996), the response (G) observed in target environment j′ due to indirect selection in test environment j is:
where i j is the selection intensity imposed in the test environment, h j and h j′ are the square roots of the narrow-sense heritability (h 2) in the test environment and the target environment, respectively, r g(jj′) is the additive genetic correlation between the test environment and the target environment, σ p(j′) is the square root of the phenotypic variance in the target environment, and σ g(j′) is the square root of the additive genetic variance in the target environment. From Eq. 1, it is clear that the usefulness of the test environment in indirect selection for the target environment has to be evaluated with regard to two aspects: (1) the heritability for the trait of interest in the environment (h 2 j ), and (2) its genetic correlation with the target environment (r g(jj′)). In the terminology of Allen et al. (1978), the proper measure of the value of a test environment is \( r\sqrt H \) where r is the correlation between genotypic performance in the test environment and the target environments and H = h 2 is the heritability in the test environment.
In addition to identifying superior genotypes, genotype-by-environment data from multi-environment trials are also valuable for evaluating the test environments (Cooper et al. 1996; Yan et al. 2007). Yan (2001) proposed that each test environment can be graphically evaluated in a GGE biplot for (1) its power to discriminate the genotypes, measured by its vector length in the biplot, and (2) its representativeness of other test environments, measured by its angle with the “average” environment. Intuitively, these two aspects should correspond to the heritability and the genetic correlation in the indirect selection theory but a clear connection between the two concepts has not been established.
A GGE biplot is a biplot (Gabriel 1971) based on environment-centered data, which removes the environment main effect and integrates the genotypic main effect with the genotype-by-environment interaction effect of a genotype-by-environment dataset (Yan et al. 2000). For a given dataset, different GGE biplots can be generated, depending on how the data are scaled prior to singular value decomposition. The most commonly used data scaling methods include (1) no scaling, (2) scaling by the standard deviation of genotype means within environments (SD-scaled), and (3) scaling by the standard error within environments (SE-scaled) (Yan et al. 2000). The SE-scaled GGE biplot has been identified as most appropriate for genotype and test environment evaluation because it accounts for heterogeneity among environments in experimental errors (Yan et al. 2000; Yan and Kang 2003; Blanche and Myers 2006). However, often a GGE biplot is used in genotype and test environment evaluation without mentioning the scaling method, which may lead to misinterpretation of the data. This occurs mainly due to the lack of understanding of the links between biplot interpretation and the indirect selection theory.
The objectives of this paper were to (1) establish mathematical connections between key parameters in the indirect selection theory and GGE biplot analysis; (2) introduce a heritability-adjusted (HA) GGE biplot, in which the expected vector length of the test environments is the square root heritability in the environments, and (3) demonstrate the use of this HA-GGE biplot in test environment evaluation and genotype evaluation using an oat (Avena sativa L.) multi-environment trial dataset.
Theory development
Common statistics characterizing the test environment
In a multi-environment trial framework, each test environment is typically characterized by the following statistical parameters relative to the trait of interest:
-
The mean, i.e., average value across genotypes in the environment. It is the environment main effect, which is not pertinent to genotype evaluation. It is not pertinent to test environment evaluation either because it does not affect the relative differences among genotypes.
-
The standard error (SE), which is the square root of the error variance in the environment (SE = σ e ), which can be reduced by various methods, including improved experimental design, improved experimental execution, and improved data analyses (Casanoves et al. 2005; Gilmour et al. 1997; Smith et al. 2002).
-
The standard deviation of genotype means (SD), which equals the square root of the phenotypic variance within the environment (σ 2 p ), with
where σ 2 g is the genotypic variance and n is the number of replicates within the environment.
-
The coefficient of variation (CV), which is the ratio of standard error over the environmental mean: CV% = (SE/Mean) × 100%. CV is sometimes used as a measure of validity of the trial and a criterion to exclude trials with low precision, although such use has been criticized (Bowman and Watson 1997; Taylor et al. 1999).
-
The heritability in the broad sense (H), which is calculated as
H is an indication of the validity or usefulness of the trial in genotype evaluation. H = 1 means that the observed differences in genotypic means in the trial are entirely due to genetic effects; H = 0 indicates that the observed differences are completely due to random error.
Basic types of GGE biplots
The general model for GGE biplots is:
where p ij represents the genotype-by-environment two-way table of GGE effects with i = 1,…,m genotypes and j = 1,…,e environments, which is decomposed into k = 1 to t principal components (PC), with t ≤ min(e, m − 1). \( \bar{y}_{ij} \) is the cell mean of genotype i in environment j; μ j is the mean value in environment j. The operation \( (\bar{y}_{ij} - \mu_{j} ) \) is referred to as “environment-centering”, which results in removal of environment main effects (E) from the original data. The model is subject to the constraint \( \lambda_{1} \ge \lambda_{2} \ge \cdots \ge \lambda_{t} \ge 0 \) and to orthonormality on the α ik scores, i.e., \( \sum\nolimits_{i = 1}^{m} {\alpha_{ik} \alpha_{ik'} } = 1 \) if k = k′ and \( \sum\nolimits_{i = 1}^{m} {\alpha_{ik} \alpha_{ik'} } = \, 0 \) if \( k \ne k' \), with similar constraints on the γ jk scores [defined by replacing symbols (i, m, α) with (j, e, γ)]. The \( \bar{\varepsilon }_{ij} \) within an environment are assumed \( NID(0,\;\sigma^{2} /n) \), where σ 2 is the experimental error variance and n is the number of replicates within an environment. A two-dimensional GGE biplot is constructed using the first two principal components (PC1 and PC2), thus t = 2 (see Yan et al. 2007 for more detailed descriptions).
Of particular relevance to this study is the parameter s j , which is referred to as the scaling factor, and the operation of division of the data matrix by s j is referred to as “data scaling”. For a given genotype-by-environment two-way table, different GGE biplots can be generated, depending on how s j is defined (Yan and Kang 2003; Yan and Tinker 2006).
Unscaled GGE biplot
It is called unscaled GGE biplot if
SE-scaled GGE biplot
It is called SE-scaled GGE biplot if
where SE j is the standard error within environment j. As a variant of the SE-scaled GGE biplot, it is called Pairwise SE-scaled GGE biplot if
which is the standard error for pairwise genotype comparisons in an environment. It is called SEM-scaled GGE biplot if
which is the standard error of means (SEM) in an environment. A further variant of the SE-scaled GGE biplot is to scale the data with the least significant difference (LSD) within each environment:
SD-scaled GGE biplot
The biplot is referred to as SD-scaled GGE biplot if
where SD j is the standard deviation of the distribution of genotype means within environment j.
Interpretation of the environmental vector length in different GGE biplots
For an unscaled GGE biplot, the vector length of environment j is
When the data are environment-centered such that the environment means \( \bar{y}_{j} \) become 0, the SD in an environment is
From Eqs. 8 and 9, it can be seen that when the data are environment-centered but not scaled (Eq. 5), the vector length of an environment is \( \sqrt {m - 1} \) times the standard deviation of genotype means within the environment (Kroonenberg 1995; Yan and Tinker 2006):
Considering the relationship between SD and H (Eq. 3), Eq. 10 can be written as
Thus, the vector length of an environment (L j ) in the unscaled GGE biplot (Eq. 5) is proportional to the phenotypic variation among genotypes (SD j , Eq. 2), which is positively associated with both the experimental error (SE j ) and the heritability (H j ) in the environment. The number of replicates n is also a factor to affect the vector length of the environment.
In the SE-scaled GGE biplot (Eq. 6), the vector length of environment j is determined solely by the heritability in the environment, though in a curvilinear fashion, given the same number of genotypes m and the same number of replications n:
The factor n can be removed from this relationship if the data are scaled by the pairwise standard error (Eq. 6a) because:
Similarly, if the data are scaled by the SEM (Eq. 6b), then:
Equations 12a and 12b provide a clear connection between the interpretation of the vector length of test environments in the GGE biplot (Eq. 8) and the heritability (H) in the theory of indirect selection (Eq. 2). That is, when all environments have the same number of genotypes (m), the vector length of the environments in the pairwise SE- or SEM-scaled GGE biplot is solely determined by, and therefore represents, the heritability or repeatability of genotype means in the environments, assuming a perfect fit of the GGE biplot. However, the relationship between the vector length and H is curvilinear rather than linear. When H is small, say less than about 0.75, the relationship appears approximately linear, but when H is large, say above about 0.95, the discriminatory power of that environment will be overstated in the GGE biplot (data not shown).
In the SD-scaled GGE biplot (Eq. 7), the vector length of all environments is a constant if the same set of genotypes are tested in all environments, since
Therefore, unlike the unscaled or SE-scaled GGE biplots, the vector length of the environments in the SD-scaled GGE biplot is not a measure of the discriminating power of the environments. Rather, all environments are expected to have the same or similar vector length if the GGE biplot adequately approximates the SD-scaled genotype-by-environment data. For the same reason, if some environments have considerately shorter vectors than others, it indicates that the SD-scaled GGE biplot does not adequately display the patterns regarding these environments. One consequence of this is that the correlations between environments with shorter vectors and other environments may not be correctly displayed by the angles between them (for an example see Yan and Frégeau-Reid 2008).
The heritability-adjusted GGE biplot
Based on Eq. 3, we have
Therefore, scaling by SEM is equivalent to scaling by \( SD\sqrt {1 - H} \), which leads to an expected vector length of the environment proportional to \( \sqrt {1 - H} \) (Eq. 12b). If the data are scaled by the factor \( SD/\sqrt H \), which is equivalent to multiplying \( \sqrt H \) in each environment to the SD-scaled data (standardized data), the expected vector length of the environments in the resulting GGE biplot would be:
Verbally, the vector length of a test environments will be proportional to the square root of the heritability in the environment, \( \sqrt H \). Such a biplot will be referred to as heritability-adjusted (HA) GGE biplot. Equation 14 establishes a strictly linear relationship between \( \sqrt H\) and the expected vector length in the HA-GGE biplot.
Interpretation of the angle between two environment vectors in different GGE biplot
When the GGE biplot has a perfect goodness of fit, the cosine of the angle (α jj′) between two environments is the genetic correlation between them (Gabriel 1971; Kroonenberg 1995; Yan and Tinker 2006):
This relationship is not altered by data scaling, as scaling with a positive value does not alter the relative differences among genotypes within an environment. Note that in the GGE biplot the target environment j′ can be an individual test environment or the “average” test environment (Yan 2001).
Test environment evaluation based on heritability-adjusted GGE biplot
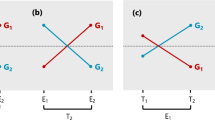
In a HA-GGE biplot, since the cosine of the angle between the vectors of two test environments (a test environment and a target environment) approximates the genetic correlation between them (Eq. 15), and since the vector length of the test environments are proportional to their \( \sqrt H \) (Eq. 14), the projection of test environment onto the target environment should approximate \( r\sqrt H \), which is an overall measure of the usefulness of a test environment (Allen et al. 1978). Thus, a HA-GGE biplot graphically displays not only r and \( \sqrt H \) but also \( r\sqrt H \). This relationship is graphically illustrated in Fig. 1.
Since the genetic gain observed in target environment j′ from indirect selection practiced in environment j is given in Eq. 1 as:
the values of h j r g(jj′) for different test environments (j) are directly proportional to the predicted indirect genetic gains in the target environment from selection in the test environments at a constant selection intensity, because σ g(j′) is a constant for a given target environment. \( r\sqrt H \) is a valid approximation of h j r g(jj′) if most genetic variation and covariation is additive, as expected for inbred lines. This may not be true for clonally propagated or full-sib families in species where dominance variation is an important component of genetic variance. In cases of predominantly additive genetic variation, however, projections of test environment vectors onto target environment vectors in a HA-GGE biplot provide a means to compare the relative utility of test environments for indirect selection for a common target environment.
Case study
A case study is presented here to demonstrate the usefulness of the HA-GGE biplot in genotype-by-environment data analysis, in comparison with the commonly used GGE biplot based on unscaled data or standardized data. The focus here is on test environment evaluation; however, as an integral part of genotype-by-environment data analysis, genotype evaluation based on the HA-GGE biplot will also be demonstrated.
Materials and methods
The data used in this case study are grain yield data of 27 covered spring oat lines and nine check cultivars tested at nine locations across eastern Canada in 2006. The locations were Bornholm (ON1), Nairn (ON2), Ottawa (ON3) and New Liskeard (ON4) in Ontario; Hebertville (QC1), Normandin (QC2), Princeville (QC3), and Amqui (QC4) in Quebec; Charlottetown in Prince Edward Island (PEI); and Hartland in New Brunswick (NB). At each location, the experimental design was a randomized complete block design. The number of replicates was four at the Ontario locations and three at other sites except QC4, where the test was not replicated (Table 1). All 27 experimental lines were tested at all sites, but the check varieties used in Ontario, Quebec, and the Maritimes (which includes New Brunswick and Prince Edward Island) were different, and no check varieties were included at ON4 and NB. The ANOVA table and basic statistics for each test environment are presented in Table 1. ANOVA was conducted based on the general linear model for randomized complete blocks and the significance of various effects was based on F test against the experimental errors within environments.
Two-dimensional un-scaled GGE biplot (Fig. 2), HA-GGE biplot (Fig. 3), and SD-scaled GGE biplot (Fig. 4) were generated using the GGEbiplot software (Yan 2001). Only the 27 breeding lines, which were tested in all environments (i.e., sites), were included in the biplots because biplots require a balanced genotype-by-environment table. All biplots in Figs. 2, 3, 4 and 5 were based on environment-focused singular value partitioning (“SVP = 2”, Yan 2002) so that they are appropriate for test environment evaluation. An environment-by-parameter biplot (Fig. 6) was also constructed to help visualize the interrelationships between basic statistics characterizing the test environments and the vector length of the environments from the various GGE biplots. Test environment evaluation and genotype evaluation are meaningful only within a mega-environment (Yan et al. 2007). Two HA-GGE biplots were constructed for the Quebec and Maritime sites to illustrate their application in test environment evaluation (Fig. 7) and genotype evaluation (Fig. 8) within a mega-environment.
Un-scaled GGE biplot. “Scaling = 0” means the data were not scaled; “Centering = 2” means the data were centered by the means of the environments. “SVP = 2” means the singular values were partitioned into the environment eigenvectors and therefore the biplot is appropriate for visualizing the correlations among the environments. For site (environment) abbreviations see Table 1
Heritability-adjusted GGE biplot. “Scaling = 4” means the heritability adjusted, which mean multiplying the heritability in each environment to the environment-standardized data; “Centering = 2” means the data were centered by the means of the environments. “SVP = 2” means the singular values were partitioned into the environment eigenvectors and therefore the biplot is appropriate for visualizing the correlations among the environments. For site (environment) abbreviations see Table 1
SD-scaled GGE biplot. “Scaling = 1” means the data were scaled by the standard deviation of genotype means within environments; “Centering = 2” means the data were centered by the means of the environments. “SVP = 2” means the singular values were partitioned into the environment eigenvectors and therefore the biplot is appropriate for visualizing the correlations among the environments. For site (environment) abbreviations see Table 1
SE-scaled GGE biplot. “Scaling = 3” means the data were scaled by the standard error (SE) of genotype means within environments; “Centering = 2” means the data were centered by the means of the environments. “SVP = 2” means the singular values were partitioned into the environment eigenvectors and therefore the biplot is appropriate for visualizing the correlations among the environments. For site (environment) abbreviations see Table 1
Environment-by-parameter biplot to show the interrelationships among parameters characterizing the test environments. “Scaling = 1” means the data were scaled by the standard deviations within parameters; “Centering = 2” means the data were centered by the means of the parameters. “SVP = 2” means the singular values were partitioned into the parameter eigenvectors and therefore the biplot is appropriate for visualizing the correlations among the parameters. CV% coefficient of variation of the environments, H-SQRT square root of the heritability of the environments, L-HA vector length of the environments in the HA-GGE biplot, L-SD vector length of the environments in the SD-scaled GGE biplot, L-LSD vector length in the LSD-scaled GGE biplot, L-US vector length of the environments in the un-scaled GGE biplot, SD standard deviation of the environments, SE standard error of the environments. For environment abbreviations see Table 1. Note that L-LSD and L-HA are heavily overlapped
Heritability-adjusted GGE biplot for Quebec and maritime sub-region. “Scaling = 2” means heritability adjusted, which means multiplying the heritability in each environment to the environment-standardized data; “Centering = 2” means the data were centered by the means of the environments. “SVP = 2” means the singular values were partitioned into the environment eigenvectors and therefore the biplot is appropriate for visualizing the correlations among the environments. The small circle represents the average environment and the thick line with an arrow pointing is the average environment vector and pointing to greater \( r\sqrt H \) value. For site (environment) abbreviations see Table 1
The mean versus stability view of the heritability-adjusted GGE biplots for the Quebec and Maritime sub-region. “Scaling = 2” means the heritability adjusted, which means multiplying the square root of heritability in each environment to the environment-standardized data; “Centering = 2” means the data were centered by the means of the environments. “SVP = 1” means the singular values were partitioned into the genotype eigenvectors and therefore the biplot is appropriate for visual comparison among genotypes. For environment abbreviations see Table 1. The single-arrowed line points to high mean performance across environments and the double-arrowed lines point to greater performance variability across environments regardless of direction
Interpretation of the environmental vector length in different GGE biplots
The vector lengths of the test environments in the un-scaled GGE biplot (Fig. 2) approximate the phenotypic variances within the environments (Eq. 11), as demonstrated by the close positioning of “SD” and “L-US” in the environment-by-parameter biplot (Fig. 6). QC3 exhibited the greatest phenotypic variance, but it also had the highest experimental error and CV (Table 1; Figs. 2, 6).
The vector length of the environments in the HA-GGE biplot (Fig. 3) approximates their square root heritabilities (Equation [12b]), as is confirmed graphically in Fig. 6, where the vector length in the HA-GGE biplot (“L-HA”) and \( \sqrt H \) had a very small angle. Figure 3 shows that ON1 and ON2 were most discriminating (with the highest \( \sqrt H \)), followed by QC1 and QC2. ON3 and ON4 were considerably less discriminating. The difference in vector length among environments was not manifest in the SE-scaled GGE biplot (Fig. 5), with clearly long vectors for ON1, ON2, QC1, and QC4 and shorter vectors for others. The correlation between environmental vector length and \( \sqrt H \) for the SE-scaled GGE biplot was even slightly higher than that for the HA-GGE biplot (Fig. 6). According to Fig. 6, the vector length of the environments in the SD-scaled GGE biplot were also, quite unexpectedly, highly correlated with \( \sqrt H \). Examination of other datasets (not reported here), however, indicates that this was merely a coincident for this particular dataset.
The unscaled GGE biplot is an adequate approximation of the HA-GGE biplot only when the SD and H are similar among environments. For this particular dataset, the phenotypic variance (SD) and the square root heritability \( \sqrt H \) were only loosely correlated as indicated by the large angle between them (Fig. 6), suggesting that the unscaled GGE biplot was not a good substitution for test environment evaluation and genotype evaluation.
The vector length of the environments in the SD-scaled GGE biplot (Fig. 4) was similar for all environments except ON3 and ON4. This is not an indication that ON3 and ON4 were less discriminating; rather, it indicates that patterns related to these two environments were not fully displayed in the biplot, which indicates that these two environments were less associated with other environments than the angles in the biplot would suggest (Yan and Frégeau-Reid 2008).
A close positive association between CV and “SE” and a loose negative association between CV and \( \sqrt H \) were observed (Fig. 6). This indicates that CV was a poor representation of H. Therefore, CV is not a good measure of the validity of the trials and should not be used in judging the validity of trials in multi-environment data analyses (Bowman and Watson 1997; Taylor et al. 1999).
Interpretation of the angles between environments in different GGE biplots
According to Eq. 15, the cosine of the angle between two environments in the GGE biplot approximates the genotypic correlation between them, regardless of the data scaling methods. Although there are no strict relations, the goodness of approximation for the correlation coefficients by the angles is related to the goodness of fit of the biplot. The un-scaled GGE biplot (Fig. 2) reveals two groups of environments: the four Ontario environments as one group and the Quebec and Maritime environments as the other. Between groups, the environments were un-associated (right angles) or negatively associated (obtuse angles). Within each group, the environments were more or less positively correlated (acute angles). Within the Quebec and Maritime group, QC3 were apparently less associated with other sites. The same statements hold for the HA (Fig. 3), the SD-scaled (Fig. 4), and the SE-scaled (Fig. 5) GGE biplots. However, the environment grouping is most clear in the SD-scaled biplot due to similar vector length among environments. This property makes the SD-scaled GGE biplot the preferred biplot if the main purpose is to investigate the associations among test environments. For simultaneous evaluation of both the discrimination power and the representativeness of the test environments, however, the HA-GGE biplot is most appropriate. Note that the angles among the test environments are not identical in the four GGE biplots (Figs. 2, 3, 4, 5), this is probably due to the fact that the biplots differ in goodness of fit. For the current dataset, the goodness of fit was 66.6% for the unscaled GGE biplot, 57.8% for the HA-scaled GGE biplot, 55.6% for the SD-scaled GGE biplot, and 72.2% for the SE-scaled GGE biplot. The interpretation of the angle between the vectors of two environments is also true for the biplot generated from Factor Analytic models (Smith et al. 2002), which may be regarded as the random effect version of the GGE biplot.
Interpretation of the angle between a test environment and the average environment
From the indirect selection theory (Eq. 1) or the formula of Allen et al. (1978), it is clear that genetic correlation (r) and the square root heritability (\( \sqrt H \)) must be considered simultaneously in assessing the usefulness of a test environment. It is a desirable feature of the HA-GGE biplot to graphically and simultaneously display both factors, both separately and jointly (Fig. 1). A test environment is not useful if its \( \sqrt H \) is very low or its genetic correlation with the target environment is small or negative. Test environment evaluation in the context of multi-environment variety trials is different in that the target environment is not a single, well defined environment. Rather, it is a population of environments that is represented (presumably) by all the test environments. To represent the target environment, Yan (2001) defined a virtual “average environment”, which is the point on the biplot that has the average coordinates of all test environments (the small circle, Fig. 7). Smith et al. (2002) similarly defined an “average environment” as a reference for selecting representative test sites based on a Factor Analytic plot of test environments.
Yan et al. (2007) proposed that the angle between a test environment and the average environment in the GGE biplot (the line that passes through the biplot origin and the average environment) is a measure of its representativeness of the target environment. Based on the discrimination power (vector length) and the correlation with the average environment, test environments can be classified into four types: (1) non-discriminating and therefore useless; (2) discriminating and representative, ideal for selecting superior genotypes; (3) discriminating but non-representative, useful for culling unstable genotypes; and (4) discriminating, but negatively correlated with the average environment, which are misleading if used in genotype selection. This classification of test environments is meaningful only when all test environments belong to a common target mega-environment (Yan et al. 2007). When strongly negative correlations exist among test environments, one should investigate whether the target environments can be divided into meaningful mega-environments.
Test environment evaluation based on the HA-GGE biplots
Figure 7 was constructed for the Quebec and Maritime sub-region to show how test environments can be evaluated based on a HA-GGEbiplot. The single-arrowed line passes through the biplot origin and the “average” environment. It represents the target environment and may be referred to as the Target Environment Axis (TEA). The projection of each test environments onto TEA approximate its \( r\sqrt H \), and are a measure of its usefulness in selecting superior genotypes for the mega-environment (Fig. 1). Thus, the usefulness of the five environments can be ranked as: QC1 > QC2 > NB > PEI > QC3 (Fig. 7). The line with two arrows points away from representativeness, regardless of directions. Thus QC3 was the least representative environment; such an environment is not good for selecting superior genotypes but can be useful for culling unstable genotypes, if it is sufficiently discriminating.
Genotype evaluation based on the HA-GGE biplots
Although the focus of this study is on test environment evaluation, the ultimate goal is to improve the efficiency of genotype evaluation. The HA-GGE biplot is the most appropriate for genotype evaluation as well as for test environment evaluation. However, the form of GGE biplot in Fig. 7 is not the best for genotype evaluation because it is based on environment-focused singular value partitioning (“SVP = 2”), which is appropriate only for evaluating test environments. Genotype-focused singular value partitioning (“SVP = 1”, Yan 2002) is required for the latter purpose (Fig. 8). For the Quebec and Maritime region, the best genotypes were “1063-8” and “1130-1” (Fig. 8). While 1063-8 was stable across environments, 1130-1 was highly variable. It is interesting to mention that 1063-8 was registered as “Dieter” for its excellent performance across years for Quebec. “1130-1” continued to perform well in 2007 and 2008 in Northern Quebec represented by QC1 ad QC2.
Discussion
Multi-year data are essential for test site evaluation
In the case study, a single year multi-site test was used for the purpose of demonstration. However, multi-year data are critical for delineating mega-environments and in selecting test locations. In practice, multi-year data are rarely balanced due to the change of genotypes and locations each year. DeLacy et al. (1996) proposed four strategies in dealing with this problem. The most convenient strategy appears to be one in which multi-site data are analyzed yearly and summarized across years. Only patterns that are consistent cross years can be used in delineating mega-environments, selecting superior test sites, and discarding non-informative and redundant test sites.
Approximation of the HA-GGE biplot by other types of GGE biplots
Prior to the development of the HA-GGE biplot, three types of GGE biplots were widely used: the unscaled GGE biplot, the SD-scaled GGE biplot, and the SE-scaled GGE biplot. Given the conclusion that HA-GGE biplot is the most desired type of GGE biplot for test environment and genotype evaluation, it is relevant to discuss its relationships with the other types of GGE biplot. From the theory development section, it is clear that the SD-scaled GGE biplot is identical to the HA-GGE biplot if all environments have the same heritability H. The unscaled GGE biplot is identical to the HA-GGE biplot if all environments have the same SD and H. Therefore, the SD-scaled GGE biplot has more chances to be a better approximation of the HA-GGE biplot than the unscaled GGE biplot (compare Figs. 2, 3, 4, 5). According to Eq. 3, scaling by SE is equivalent to scaling by \( SD\sqrt {n(1 - H)} \) or its variants. Thus the SE-scaled GGE biplot uses the same information as the HA-GGE biplot but in a different way. The consequence is that while there is a linear relationship between the expected environment vector length and \( \sqrt H \) in the HA-GGE biplot, this relationship for the SE-scaled GGE biplot is curvilinear. If the H values of the environments are within a reasonable range, the difference between the two types of GGE biplot should be quite small. In fact, the empirical correlation between the environmental vector length and the \( \sqrt H \) was higher for the SE-scaled GGE biplot than for the HA-GGE biplot for the current dataset and two other datasets not reported here, an observation we don’t fully understand. Therefore, although the HA-GGE biplot has a more direct link to the theory of indirect selection, conclusions derived from SE-scaled GGE biplot, such as those reported by Blanche and Myers (2006), should remain largely relevant.
GGE biplot versus scatter plot
Gauch et al. (2008) criticized the use of GGE biplot in test environment evaluation and genotype evaluation as described in Yan et al. (2007). For test environment evaluation, they presented a scatter plot of discriminating power versus representativeness and argued that the scatter plot was superior to the GGE biplot approach because the values plotted were exact values rather than approximations and the process was simpler. Similarly, for genotype evaluation, Gauch et al. (2008) presented a scatter plot of mean versus stability and argued against the GGE biplot approach. We agree that scatter plot approach are valid alternatives of the GGE biplot approach for test environment evaluation and genotype evaluation, we also agree that GGE biplots are approximate displays of the data while the scatter plots display some “exact” summaries of the data. However, GGE biplots convey information that cannot be gleaned from these scatter plots due to their simultaneous display of both genotypes and environments and their inner-product property (Gabriel 1971). GGE biplots not only display the quantities that are used in test environment and genotype evaluation but also preserves the original information from which the quantities are derived. For example, Fig. 6 demonstrates that QC3 was a poor test environment; this was partially due to the low yield of genotypes 1130-1 and 1168-3 at that site. Similarly, Fig. 8 shows that 1130-1 was a high yielding but unstable genotype resulting from its good adaptation to northern Quebec (QC1 and QC2) but poor adaptation to central Quebec (QC3). Such information is not conveyed in the scatter plots.
Single quantity versus two quantities for test environment evaluation
Gauch et al. (2008) also criticized the GGE biplot approach to test environment evaluation by citing Cooper et al. (2006): “The optimal site maximizes the phenotypic correlation between entry yields on the site and entry yields over the target environments (as represented by the test sites for a given mega-environment) because the phenotypic correlation includes the genetic correlation and the broad-sense heritability of the sites.” The HA-GGE biplot proposed in this paper accomplishes precisely this (Figs. 1, 7) because the phenotypic correlation between environments j and j′ is \( r_{p(jj')} = h_{j} h_{j'} r_{g(jj')} \) (Cooper and DeLacy 1994). The HA-GGE biplot displays h j and h j′ as the lengths of the two environment vectors and r g(jj′) by the cosine of the angle between the two vectors. This is additional to the fact that the HA-GGE biplot (Fig. 7) graphically displays r, \( \sqrt H \), and \( r\sqrt H \) for each test environment, which allows test environment evaluation based on r and\( \sqrt H \) both jointly and separately.
Conclusions
We demonstrated both theoretically and empirically that the HA-GGE biplot graphically displays the square root heritability of each test environment, \( \sqrt H \), and its genetic correlation with other test environments, r, which are the two key elements for test environment evaluation in the framework of indirect selection theory. Moreover, the HA-GGE biplot allows test environments to be ranked graphically based on their \( r\sqrt H \) values with respect to a common target environment. Therefore, the HA-GGE biplot appears to be most appropriate of all GGE biplots for visual evaluation of the test environments. This biplot is also most appropriate for genotype evaluation, because it takes into consideration any heterogeneity among environments by giving weights to the test environments proportional to their \( \sqrt H \). The SE-scaled GGE biplot is a good approximation of the HA-GGE biplot when the H values of the environments are within a reasonably small range; the SD-scaled GGE biplot is a good approximation when the environments have the same or similar H values; and the unscaled GGE biplot is a good approximation only when all environments are similar in both SD and H values.
Abbreviations
- H:
-
Heritability or repeatability of genotypic differences within an environment
- HA:
-
Heritability-adjusted
- GGE:
-
Genotype main effect plus genotype-by-environment interaction
- SD:
-
Standard deviation of genotype means within an environment
- SE:
-
Standard error within an environment
- SEM:
-
Standard error of means within an environment
References
Allen FL, Comstock RE, Rasmusson DC (1978) Optimal environments for yield testing. Crop Sci 18:747–751
Blanche SB, Myers GO (2006) Identifying discriminating locations for cultivar selection in Louisiana. Crop Sci 46:946–949
Bowman DT, Watson CE (1997) Measures of validity in cultivar performance trials. Agron J 89:860–866
Casanoves F, Macchiavelli R, Balzarini M (2005) Error variation in multienvironment peanut trials. Crop Sci 45:1927–1933
Cooper M, DeLacy IH (1994) Relationships among analytical methods used to study genotypic variation and genotype-by-environment interaction in plant breeding multi-environment experiments. Theor Appl Genet 88:561–572
Cooper M, DeLacy IH, Basford KE (1996) Relationships among analytical methods used to analyse genotypic adaptation in multi-environment trials. In: Cooper M, Hammer GL (eds) Plant adaptation and crop improvement. CAB International, Wallingford, UK, pp 193–224
DeLacy IH, Basford KE, Cooper M, Fox PN (1996) Retrospective analysis of historical data sets from multi-environment trials—theoretical development. In: Cooper M, Hammer GL (eds) Plant adaptation and crop improvement. CAB International, Wallingford, UK, pp 243–267
Falconer DS, Mackay TFC (1996) Introduction to quantitative genetics, 4th edn. Longman Scientific and Technical, Harlow, Essex
Gabriel KR (1971) The biplot graphic display of matrices with application to principal component analysis. Biometrika 58:453–467
Gauch HG (2006) Statistical analysis of yield trials by AMMI and GGE. Crop Sci 46:1488–1500
Gauch HG, Piepho H-P, Annicchiarico P (2008) Statistical analysis of yield trials by AMMI and GGE: further considerations. Crop Sci 48:866–889
Gilmour AR, Cullis BR, Verbyla AP (1997) Accounting for natural and extraneous variation in the analysis of field experiments. J Agric Biol Environ Stat 2:269–293
Guillen-Portal FR, Russell WK, Eskridge KM, Baltensperger DD, Nelson LA, D’Croz-Mason NE, Johnson BE (2004) Selection Environments for maize in the U.S. western high plains. Crop Sci 44:1519–1526
Kroonenberg PM (1995) Introduction to biplots for G×E tables. Department of Mathematics, Research Report 51. University of Queensland, Australia
Smith AB, Cullis BR, Thompson R (2002) Exploring variety—environment data using random effects AMMI models with adjustments for spatial field trend: Part I. Theory. In: Kang M (ed) Quantitative genetics, genomics, and plant breeding. CAB Int, Oxford, UK, pp 323–336
Taylor SL, Payton ME, Raun WR (1999) Relationship between mean yield, coefficient of variation, mean square error and plot size in wheat field experiments. Commun Soil Sci Plant Anal 30:1439–1447
Yan W (2001) GGEbiplot—a Windows application for graphical analysis of multi-environment trial data and other types of two-way data. Agron J 93:1111–1118
Yan W (2002) Singular value partitioning for biplot analysis of multi-environment trial data. Agron J 94(5):990–996
Yan W, Frégeau-Reid JA (2008) Breeding line selection based on multiple traits. Crop Sci 48:417–423
Yan W, Kang MS (2003) GGE biplot analysis: a graphical tool for breeders, geneticists, and agronomists. CRC Press, Boca Raton, FL
Yan W, Tinker NA (2006) Biplot analysis of multi-environment trial data: principles and applications. Can J Plant Sci 86:623–645
Yan W, Hunt LA, Sheng Q, Szlavnics Z (2000) Cultivar evaluation and mega-environment investigation based on GGE biplot. Crop Sci 40:597–605
Yan W, Kang MS, Ma B-L, Woods S, Cornelius PL (2007) GGE biplot vs. AMMI analysis of genotype-by-environment data. Crop Sci 47:643–653
Acknowledgment
We thank Judith Fregeau-Reid, Brad de Haan, Mark Etienne, John Rowsell, Richard Martin, Allan Cummiskey, Peter Scott, Denis Pageau, and Julie Durand for planning and conducting the 2006 oat yield trials at various research stations.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Yan, W., Holland, J.B. A heritability-adjusted GGE biplot for test environment evaluation. Euphytica 171, 355–369 (2010). https://doi.org/10.1007/s10681-009-0030-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-009-0030-5