Abstract
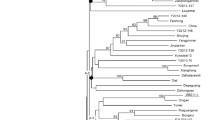
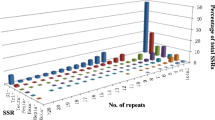
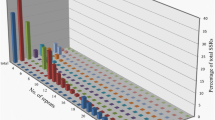
Simple sequence repeat motifs are abundant in plant genomes and are commonly used molecular markers in plant breeding. In tomato, currently available genetic maps possess a limited number of simple sequence repeat (SSR) markers that are not evenly distributed in the genome. This situation warrants the need for more SSRs in genomic regions lacking adequate markers. The objective of the study was to develop SSR markers pertaining to chromosome 6 from bacterial artificial chromosome (BAC) sequences available at Solanaceae Genomics Network. A total of 54 SSR primer pairs from 17 BAC clones on chromosome 6 were designed and validated. Polymorphism of these loci was evaluated in a panel of 16 genotypes comprising of Solanum lycopersicum and its wild relatives. Genetic diversity analysis based on these markers could distinguish genotypes at species level. Twenty-one SSR markers derived from 13 BAC clones were polymorphic between two closely related tomato accessions, West Virginia 700 and Hawaii 7996 and were mapped using a recombinant inbred line population derived from a cross between these two accessions. The markers were distributed throughout the chromosome spanning a total length of 117.6 cM following the order of the original BAC clones. A major QTL associated with resistance to bacterial wilt was mapped on chromosome 6 at similar location of the reported Bwr-6 locus. These chromosome 6-specific SSR markers developed in this study are useful tools for cultivar identification, genetic diversity analysis and genetic mapping in tomato.



Similar content being viewed by others
References
Ammiraju JSS, Veremis JC, Huang X, Roberts PA, Kaloshian I (2003) The heat-stable root-knot nematode resistance gene Mi-9 from Lycopersicon peruvianum is localized on the short arm of chromosome 6. Theor Appl Genet 106(3):478–484
Areshchenkova T, Ganal MW (1999) Long tomato microsatellites are predominantly associated with centromeric regions. Genome 42:536–544
Areshchenkova T, Ganal MW (2002) Comparative analysis of polymorphism and chromosomal location of tomato microsatellite markers isolated from different sources. Theor Appl Genet 104:229–235
AVRDC (2007) Tomato trials for the APSA workshop. In: AVRDC report 2004. AVRDC—The World Vegetable Center, Shanhua, p 32
Bai Y, Huang CC, van der Hulst R, Meijer-Dekens F, Bonnema G, Lindhout P (2003) QTLs for tomato powdery mildew resistance (Oidium lycopersici) in Lycopersicon parviflorum G1.1601 co-localize with two qualitative powdery mildew resistance genes. Mol Plant Microbe Interact 16(2):169–176
Bell CJ, Ecker JR (1994) Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19:137–144
Bernacchi D, Tanksley SD (1997) An interspecific backcross of Lycopersicon esculentum × L. hirsutum: linkage analysis and a QTL study of sexual compatibility factors and floral traits. Genetics 147:861–877
Bredemeijer GMM, Arens P, Wouters D (1998) The use of semiautomated fluorescent microsatellite analysis for tomato cultivar identification. Theor Appl Genet 97:584–590
Carmeille A, Caranta C, Dintinger J, Prior P, Luisetti J, Besse P (2006) Identification of QTLs for Ralstonia solanacearum race 3-phylotype II resistance in tomato. Theor Appl Genet 113:110–121
Chen FQ, Foolad MR (1999) A molecular linkage map of tomato based on a cross between Lycopersicon esculentum and L. pimpinellifolium and its comparison with other molecular maps of tomato. Genome 42:94–103
Edwards JD, McCouch SR (2007) Molecular markers for use in plant molecular breeding and germplasm evaluation. In: Guimarães EP, Ruane J, Scherf BD, Sonnino A, Dargie JD (eds) Marker-assisted selection. Current status and future perspectives in crops, livestock, forestry, and fish. Food and Agriculture Organization of the United Nations, Rome, pp 29–49
Foolad MR (2007) Genome mapping and molecular breeding of tomato. Int J Plant Genomics. doi:10.1155/2007/64358
Frary A, Xu Y, Liu J, Mitchell S, Tedeschi E, Tanksley SD (2005) Development of a set of PCR-based anchor markers encompassing the tomato genome and evaluation of their usefulness for genetics and breeding experiments. Theor Appl Genet 111(2):291–312
Fulton TM, Nelson JC, Tanksley SD (1997) Introgression and DNA marker analysis of Lycopersicon peruvianum, a wild relative of the cultivated tomato, L. esculentum, followed through three successive backcross generations. Theor Appl Genet 95:895–902
Fulton TM, Van der Hoeven R, Eannetta NT, Tanksley SD (2002) Identification, analysis, and utilization of conserved ortholog set markers for comparative genomics in higher plants. Plant Cell 14:1457–1467
Grandillo S, Tanksley SD (1996) Genetic analysis of RFLPs, GATA microsatelllites and RAPDs in a cross between L. esculentum and L. pimpinelliflium. Theor Appl Genet 92:957–965
Hanson PM, Bernacchi D, Green S (2000) Mapping a wild tomato introgression associated with tomato yellow leaf curl virus resistance in a cultivated tomato line. J Am Soc Hortic Sci 125(1):15–20
Hanson PM, Chen J, Kuo G (2002) Gene action and heritability of high temperature fruit set in tomato line CL5915. HortScience 37(1):172–175
He C, Poysa V, Yu K (2003) Development and characterization of simple sequence repeat (SSR) markers and their use in determining relationships among Lycopersicon esculentum cultivars. Theor Appl Genet 106:363–373
Jones DA, Dickinson MJ, Balint-Kurti PJ, Dixon MS, Jones JDG (1993) Two complex resistance loci revealed in tomato by classical and RFLP mapping of Cf-2, Cf-4, Cf-5, and Cf-9 genes for resistance to Cladosporium fulvum. Mol Plant Microbe Interact 6:348–357
Kosambi D (1944) The estimation of map distances from recombination values. Ann Eugen 12:172–175
Kumpatla SP, Mukhopadhyay S (2005) Mining and survey of simple sequence repeats in expressed sequence tags of dicotyledonous species. Genome 48:985–998
Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimental and natural population. Genomics 1:174–181
Langella O (1999) Populations 1.2.30. http://bioinformatics.org/~tryphon/populations/
Milbourne D, Meyer RC, Collins AJ, Ramsay LD, Gebhardt C, Waugh R (1998) Isolation, characterization and mapping of simple sequence repeat loci in potato. Mol Gen Genet 259:233–245
Moreau P, Thoquet P, Olivier J, Laterrot H, Grimsley N (1998) Genetic Mapping of Ph-2, a single locus controlling partial resistance to Phytophthora infestans in tomato. Mol Plant Microbe Interact 11(4):259–269
Morgante M, Olivieri AM (1993) PCR-amplified microsatellites as markers in plant genetics. Plant J 3:175–182
Mueller LA, Lankhorst RK, Tanksley SD, Giovannoni JJ et al (2009) A snapshot of the emerging tomato genome sequence. Plant Genome 2:78–92. doi:10.3835/plantgenome2008.08.0005
Nagy I, Polley A, Ganal M (1998) Development and characterization of microsatellite markers in pepper. In: Xth EUCARPIA meeting on genetics and breeding on Capsicum & Eggplant, Avignon, France
Nei M (1973) Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci USA 70:3321–3323
Ohyama A, Asamizu E, Negoro S, Miyatake K, Yamaguchi H, Tabata S, Fukuoka H (2009) Characterization of tomato SSR markers developed using BAC-end and cDNA sequences from genome database. Mol Breed 23:685–691
Page RDM (1996) TreeView: an application to display phylogenetic trees on personal computer. Bioinformatics 12:357–358
Paran I, Goldman I, Tanksley S, Zamir D (1995) Recombinant inbred lines for genetic mapping in tomato. Theor Appl Genet 90:542–548
Portis E, Nagy I, Sasvari Z, Stagel A, Barchi L, Lanteri S (2007) The design of Capsicum spp. SSR assays via analysis of in silico DNA sequence, and their potential utility for genetic mapping. Plant Sci 172:640–648
Sandbrink JM, van Ooijen JW, Purimahua CC, Vrielink M, Verkerk R, Zabel P, Lindhout P (1995) Localization of genes for bacterial canker resistance in Lycopersicon peruvianum using RFLPs. Theor Appl Genet 90:444–450
Sanwen H, Baoxi Z, Milbourne D, Cardle L, Guimei Y, Jiazhen G (2000) Development of pepper SSR markers from sequence databases. Euphytica 117:163–167
Sharma A, Zhang L, Nino-Liu D, Ashrafi H, Foolad MR (2008) A Solanum lycopersicum × Solanum pimpinellifolium linkage map of tomato displaying genomic locations of R-Genes, RGAs, and candidate resistance/defense-response ESTs. Int J Plant Genomics. doi:10.1155/2008/926090
Smulders MJ, Bredemeijer G, Rus-Kortekaas W, Arens P, Vosman B (1997) Use of short microsatellites from database sequences to generate polymorphisms among Lycopersicon esculentum cultivars and accessions of other Lycopersicon species. Theor Appl Genet 97:264–272
Srinivasa Rao NK, Bhatt RM, Sadashiva AT (2000) Tolerance to water stress in tomato cultivars. Photosynthetica 38(3):465–467
Tanksley SD, Ganal MW, Prince JP, De Vicente MC, Bonierbale MW, Broun P, Fulton TM, Giovannoni JJ, Grandillo S, Martin GB, Messeguer R, Miller JC, Miller L, Paterson AH, Pineda O, Roder MS, Wing RA, Wu W, Young ND (1992) High-density molecular linkage maps of the tomato and potato genomes. Genetics 132:1141–1160
Thoquet P, Olivier J, Sperisen C, Rogowsky P, Laterrot H, Grimsley N (1996) Quantitative trait loci determining resistance to bacterial wilt in tomato cultivar Hawaii7996. Mol Plant Microbe Interact 9(9):826–836
Van Heusden AW, Koornneef M, Voorrips RE, Brüggemann W, Pet P, Van Vrielink-Ginkel RV, Chen X, Lindhout P (1999) Three QTLs from Lycopersicon peruvianum confer a high level of resistance to Clavibacter michiganensis ssp. michiganensis. Theor Appl Genet 99:1068–1074
Wang JF, Olivier J, Thoquet P, Mangin B, Sauviac L, Grimsley NH (2000) Resistance of tomato line Hawaii7996 to Ralstonia solanacearum Pss4 in Taiwan is controlled mainly by a major strain-specific locus. Mol Plant Microbe Interact 13:6–13
Wang S, Basten CJ, Zeng ZB (2007) Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh, NC. http://statgen.ncsu.edu/qtlcart/WQTLCart.htm
Wann EV (1997) Tomato germplasm lines T4065, T4099, T5019, and T5020 with unique genotypes that enhance fruit quality. HortScience 32(4):747–748
Weber JL (1990) Informativeness of human (dC-dA)n, (dG-dT)n polymorphisms. Genomics 7:524–530
Yu K, Park SJ, Poysa V, Gebts P (2000) Integration of simple sequence repeat (SSR) markers into a molecular linkage map of common bean (Phaseolus vulgaris L.). J Hered 91(6):429–434
Zamir D, Ekstein-Michelson I, Zakay Y (1994) Mapping and introgression of a tomato yellow leaf curl virus tolerance gene, Ty-1. Theor Appl Genet 88(2):141–146
Zane L, Bargelloni L, Patarnello T (2002) Strategies for microsatellite isolation: a review. Mol Ecol 11:1–16
Zhang LP, Khan A, Niño-Liu D, Foolad MR (2002) A molecular linkage map of tomato displaying chromosomal locations of resistance gene analogs based on a Lycopersicon esculentum × Lycopersicon hirsutum cross. Genome 45(1):133–146
Acknowledgements
The authors would like to thank Drs. Kadirvel Palchamy and Robert de la Peña for providing DNA samples of the tomato genotypes used in the study, Dr. Peter Hanson for providing information on tomato accessions, and Ms. Chiou-fen Hsu for technical assistance. This study is supported by funding provided by GTZ 81070160: Deutsche Gesellshaft fur Technishe Zusammenarbeit GmbH of Germany.
Author information
Authors and Affiliations
Corresponding author
Additional information
Subramaniam Geethanjali and Kai-Yi Chen contributed equally for this manuscript.
Rights and permissions
About this article
Cite this article
Geethanjali, S., Chen, KY., Pastrana, D.V. et al. Development and characterization of tomato SSR markers from genomic sequences of anchored BAC clones on chromosome 6. Euphytica 173, 85–97 (2010). https://doi.org/10.1007/s10681-010-0125-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10681-010-0125-z