Abstract
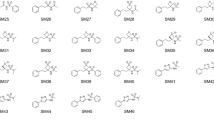
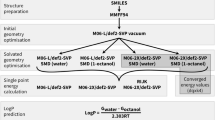
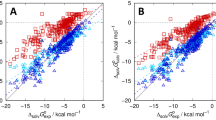
The prediction of \(\log P\) values is one part of the statistical assessment of the modeling of proteins and ligands (SAMPL) blind challenges. Here, we use a molecular graph representation method called Geometric Scattering for Graphs (GSG) to transform atomic attributes to molecular features. The atomic attributes used here are parameters from classical molecular force fields including partial charges and Lennard–Jones interaction parameters. The molecular features from GSG are used as inputs to neural networks that are trained using a “master” dataset comprised of over 41,000 unique \(\log P\) values. The specific molecular targets in the SAMPL7 \(\log P\) prediction challenge were unique in that they all contained a sulfonyl moeity. This motivated a set of ClassicalGSG submissions where predictors were trained on different subsets of the master dataset that are filtered according to chemical types and/or the presence of the sulfonyl moeity. We find that our ranked prediction obtained 5th place with an RMSE of 0.77 \(\log P\) units and an MAE of 0.62, while one of our non-ranked predictions achieved first place among all submissions with an RMSE of 0.55 and an MAE of 0.44. After the conclusion of the challenge we also examined the performance of open-source force field parameters that allow for an end-to-end \(\log P\) predictor model: General AMBER Force Field (GAFF), Universal Force Field (UFF), Merck Molecular Force Field 94 (MMFF94) and Ghemical. We find that ClassicalGSG models trained with atomic attributes from MMFF94 can yield more accurate predictions compared to those trained with CGenFF atomic attributes.






Similar content being viewed by others
References
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (1997) Adv Drug Deliv Rev 23(1–3):3
Noble A (1993) J Chromatogr A 642(1–2):3
Paschke A, Neitzel PL, Walther W, Schüürmann G (2004) J Chem Eng Data 49(6):1639
Sicbaldi F, Del Re AA (1993) Reviews of environmental contamination and toxicology. Springer, Berlin, pp 59–93
Kajiya K, Ichiba M, Kuwabara M, Kumazawa S, Nakayama T (2001) Biosci Biotechnol Biochem 65(5):1227
Hermens JL, de Bruijn JH, Brooke DN (2013) Environ Toxicol Chem 32(4):732
Schwarzenbach RP, Gschwend PM, Imboden DM (2005) Environmental organic chemistry. Wiley, New York
Cheng T, Zhao Y, Li X, Lin F, Xu Y, Zhang X, Li Y, Wang R, Lai L (2007) J Chem Inf Model 47(6):2140
Ghose AK, Crippen GM (1986) J Comput Chem 7(4):565
Leo AJ (1993) Chem Rev 93(4):1281
Meylan WM, Howard PH (1995) J Pharm Sci 84(1):83
Plante J, Werner S (2018) J Cheminf 10(1):61
Molnár L, Keserű GM, Papp Á, Gulyás Z, Darvas F (2004) Bioorg Med Chem Lett 14(4):851
Huuskonen JJ, Livingstone DJ, Tetko IV (2000) J Chem Inf Comput Sci 40(4):947
Moriguchi I, Hirono S, Liu Q, Nakagome I, Matsushita Y (1992) Chem Pharm Bull 40(1):127
Chen D, Wang Q, Li Y, Li Y, Zhou H, Fan Y (2020) Chemosphere 247:125869
Mannhold R, Poda GI, Ostermann C, Tetko IV (2009) J Pharm Sci 98(3):861
Tetko IV, Tanchuk VY, Villa AE (2001) J Chem Inf Comput Sci 41(5):1407
ADMET Predictor(TM) version 2.3.0, Simulations Plus, Inc
CSLogP version 2.2.0.0, ChemSilico LLC, USA, http://www.chemsilico.com
Silicos-it, Filter-it version 1.0.2, http://silicos-it.be.s3-website-eu-west-1.amazonaws.com/software/filter-it/1.0.2/filter-it.html
Wu K, Zhao Z, Wang R, Wei GW (2018) J Comput Chem 39(20):1444
Korshunova M, Ginsburg B, Tropsha A, Isayev O (2021) J Chem Inf Model 61(1):7
Donyapour N, Hirn M, Dickson A (2021) J Comput Chem 42(14):1006
SAMPL challenges, http://samplchallenges.github.io
Işık M, Bergazin TD, Fox T, Rizzi A, Chodera JD, Mobley DL (2020) J Comput Aid Mol Des 34(4):335–370
Bergazin TD, Tielker N, Zhang Y, Mao J, Gunner MR, Ballatore C, Kast S, Mobley D et al (2021) ChemRxiv. https://doi.org/10.26434/chemrxiv.14461962.v1
Popova M, Isayev O, Tropsha A (2018) Sci Adv 4(7):7885
Lui R, Guan D, Matthews S (2020) J Comput Aid Mol Des 34:523
Krämer A, Hudson PS, Jones MR, Brooks BR (2020) J Comput Aid Mol Des 32:983
Ding Y, Xu Y, Qian C, Chen J, Zhu J, Huang H, Shi Y, Huang J (2020) J Comput Aid Mol Des 298:31
Riquelme M, Vöhringer-Martinez E (2020) J Comput Aid Mol Des 34(1):39–54
Fan S, Iorga BI, Beckstein O (2020) J Comput Aid Mol Des 30:1045
Procacci P, Guarnieri G (2019) J Comput Aid Mol Des 35:49–61
Marenich AV, Cramer CJ, Truhlar DG (2009) J Phys Chem B 113(18):6378
Loschen C, Reinisch J, Klamt A (2020) J Comput Aid Mol Des 34(4):385
Tielker N, Tomazic D, Eberlein L, Güssregen S, Kast SM (2020) J Comput Aid Mol Des 34:709–715
Guan D, Lui R, Matthews S (2020) J Comput Aid Mol Des 34:535
Jones MR, Brooks BR (2020) J Comput Aid Mol Des 34:535
Ouimet JA, Paluch AS (2020) J Comput Aid Mol Des 34:574
Zamora WJ, Pinheiro S, German K, Ràfols C, Curutchet C, Luque FJ (2020) J Compu Aid Mol Des 34(4):443
Wang S, Riniker S (2019) J Comput Aid Mol Des 34:393
Patel P, Kuntz DM, Jones MR, Brooks BR, Wilson AK (2020) J Comput Aid Mol Des 34:495
Arslan E, Findik BK, Aviyente V (2020) J Comput Aid Mol Des 34:463
Port A, Bordas M, Enrech R, Pascual R, Rosés M, Ràfols C, Subirats X, Bosch E (2018) Eur J Pharm Sci 122:331
NonStar, logP database, https://ochem.eu/article/17434
Ma J, Sheridan RP, Liaw A, Dahl GE, Svetnik V (2015) J Chem Inf Model 55(2):263
Lusci A, Pollastri G, Baldi P (2013) J Chem Inf Model 53(7):1563
Feinberg EN, Sur D, Wu Z, Husic BE, Mai H, Li Y, Sun S, Yang J, Ramsundar B, Pande VS (2018) ACS Cent Sci 4(11):1520
Gao P, Zhang J, Sun Y, Yu J (2020) Phys Chem Chem Phys 22(41):23766
Duvenaud DK, Maclaurin D, Iparraguirre J, Bombarell R, Hirzel T, Aspuru-Guzik A, Adams RP (2015) Adv Neural Inf Process Syst 28:2224–2232
Smith JS, Isayev O, Roitberg AE (2017) Chem Sci 8(4):3192
Gao F, Wolf G, Hirn M (2019) International conference on machine learning, pages 2122–2131
Vanommeslaeghe K, MacKerell AD Jr (2012) J Chem Inf Model 52(12):3144
Vanommeslaeghe K, Raman EP, MacKerell AD Jr (2012) J Chem Inf Model 52(12):3155
Maier JA, Martinez C, Kasavajhala K, Wickstrom L, Hauser KE, Simmerling C (2015) J Chem Theory Comput 11(8):3696
Vassetti D, Pagliai M, Procacci P (2019) J Chem Theory Comput 15(3):1983
Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA (2004) J Comput Chem 25(9):1157
Rappé AK, Casewit CJ, Colwell K, Goddard WA III, Skiff WM (1992) J Am Chem Soc 114(25):10024
Halgren TA (1996) J Comput Chem 17(5–6):490
Halgren TA (1996) J Comput Chem 17(5–6):520
Hassinen T, Peräkylä M (2001) J Comput Chem 22(12):1229
Francisco KR, Varricchio C, Paniak TJ, Kozlowski MC, Brancale A, Ballatore C (2021) Eur J Med Chem 218:113399
RDkit, Open-source cheminformatics, https://www.rdkit.org
Howard P, Meylan W (1999) Physical/chemical property database (PHYSPROP), Syracuse Research Corp, Environmental Science Center, North Syracuse, NY, 1999. http://www.syrres.com/esc/physdemo.htm
Huuskonen JJ, Villa AE, Tetko IV (1999) J Pharm Sci 88(2):229
Klopman G, Li JY, Wang S, Dimayuga M (1994) J Chem Inf Comput Sci 34(4):752
Hansch C, Leo A, Hoekman D (1995) Exploring QSAR: Fundamentals and Applications in Chemistry and Biology, American Chemical Society, Washington, DC
O’Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR (2011) J Cheminf 3(1):33
The Open babel package, version 3.1.1, http://openbabel.org
Kipf TN, Welling M (2016) arXiv preprint arXiv:1609.02907
Paszke A, Gross S, Massa F, Lerer A, Bradbury J, Chanan G, Killeen T, Lin Z, Gimelshein N, Antiga L et al (2019) Adv Neural Inf Process Syst 32:8024–8035
Tietz M, Fan TJ, Nouri D, Bossan B (2017) skorch Developers, skorch: A scikit-learn compatible neural network library that wraps PyTorch. https://skorch.readthedocs.io/en/stable/
Kingma DP, Ba J (2014) arXiv preprint arXiv:1412.6980
Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V et al (2011) J Mach Learn Res 12:2825
Heskes T, Wiegerinck W, Kappen H (1997) Prog Neural Process 375:128–135
Kumar S, Srivastava A (2012) Proceedings on 18th ACM SIGKDD conference knowledgement discovery data mining
Nix DA, Weigend AS (1994) in Proceedings of 1994 ieee international conference on neural networks (ICNN’94), vol. 1 (IEEE, 1994), vol. 1, pp. 55–60
Author information
Authors and Affiliations
Corresponding author
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Donyapour, N., Dickson, A. Predicting partition coefficients for the SAMPL7 physical property challenge using the ClassicalGSG method. J Comput Aided Mol Des 35, 819–830 (2021). https://doi.org/10.1007/s10822-021-00400-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-021-00400-x