Abstract
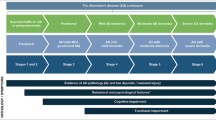
Alzheimer’s disease (AD) is an incurable neurodegenerative disorder accounting for 70%–80% dementia cases worldwide. Although, research on AD has increased in recent years, however, the complexity associated with brain structure and functions makes the early diagnosis of this disease a challenging task. Resting-state functional magnetic resonance imaging (rs-fMRI) is a neuroimaging technology that has been widely used to study the pathogenesis of neurodegenerative diseases. In literature, the computer-aided diagnosis of AD is limited to binary classification or diagnosis of AD and MCI stages. However, its applicability to diagnose multiple progressive stages of AD is relatively under-studied. This study explores the effectiveness of rs-fMRI for multi-class classification of AD and its associated stages including CN, SMC, EMCI, MCI, LMCI, and AD. A longitudinal cohort of resting-state fMRI of 138 subjects (25 CN, 25 SMC, 25 EMCI, 25 LMCI, 13 MCI, and 25 AD) from Alzheimer’s Disease Neuroimaging Initiative (ADNI) is studied. To provide a better insight into deep learning approaches and their applications to AD classification, we investigate ResNet-18 architecture in detail. We consider the training of the network from scratch by using single-channel input as well as performed transfer learning with and without fine-tuning using an extended network architecture. We experimented with residual neural networks to perform AD classification task and compared it with former research in this domain. The performance of the models is evaluated using precision, recall, f1-measure, AUC and ROC curves. We found that our networks were able to significantly classify the subjects. We achieved improved results with our fine-tuned model for all the AD stages with an accuracy of 100%, 96.85%, 97.38%, 97.43%, 97.40% and 98.01% for CN, SMC, EMCI, LMCI, MCI, and AD respectively. However, in terms of overall performance, we achieved state-of-the-art results with an average accuracy of 97.92% and 97.88% for off-the-shelf and fine-tuned models respectively. The Analysis of results indicate that classification and prediction of neurodegenerative brain disorders such as AD using functional magnetic resonance imaging and advanced deep learning methods is promising for clinical decision making and have the potential to assist in early diagnosis of AD and its associated stages.










Similar content being viewed by others
References
Selkoe, D. J., and Hardy, J., The amyloid hypothesis of Alzheimer’s disease at 25 years. EMBO Mol. Med. 8(6):595–608, Jun. 2016.
Qiang, W., Yau, W.-M., Lu, J.-X., Collinge, J., and Tycko, R., Structural variation in amyloid-β fibrils from Alzheimer’s disease clinical subtypes. Nature 541(7636):217–221, Jan. 2017.
Eftekharzadeh, B., Daigle, J. G., Kapinos, L. E., Coyne, A., Schiantarelli, J., Carlomagno, Y., Cook, C., Miller, S. J., Dujardin, S., Amaral, A. S., Grima, J. C., Bennett, R. E., Tepper, K., DeTure, M., Vanderburg, C. R., Corjuc, B. T., DeVos, S. L., Gonzalez, J. A., Chew, J. et al., Tau Protein Disrupts Nucleocytoplasmic Transport in Alzheimer’s Disease. Neuron 99(5):925–940.e7, Sep. 2018.
Khan, S. S., Bloom, G. S., Tau: The Center of a Signaling Nexus in Alzheimer’s Disease, Front. Neurosci., vol. 10, Feb. 2016.
Bachstetter, A. D., Van Eldik, L. J., Schmitt, F. A., Neltner, J. H., Ighodaro, E. T., Webster, S. J., Patel, E., Abner, E. L., Kryscio, R. J., and Nelson, P. T., Disease-related microglia heterogeneity in the hippocampus of Alzheimer’s disease, dementia with Lewy bodies, and hippocampal sclerosis of aging. Acta Neuropathol. Commun. 3(1):32, Dec. 2015.
Prince, M. J., World Alzheimer report 2015: The global impact of dementia: An analysis of prevalence, incidence, cost and trends. Alzheimer’s Disease International, 2015.
“2018 Alzheimer’s disease facts and figures,” Alzheimer’s Dement., vol. 14, no. 3, pp. 367–429, Mar. 2018.
“2018 ALZHEIMER’S DISEASE FACTS AND FIGURES Includes a Special Report on the Financial and Personal Benefits of Early Diagnosis.”
“ADNI | Background & Rationale.” .
Montgomery, V., Stabler, A., Harris, K., and Lu, L., B-26 Effects of Delay Duration on the Wechsler Memory Scale Logical Memory Performance of Older Adults with Probable Alzheimer’s Dementia, Probable Vascular Dementia, and Normal Cognition. Arch. Clin. Neuropsychol. 30(6):531, 2015.
Wyss-Coray, T., Ageing, neurodegeneration and brain rejuvenation. Nature 539(7628):180, 2016.
A. Association and others, “2017 Alzheimer’s disease facts and figures,” Alzheimer’s Dement., vol. 13, no. 4, pp. 325–373, 2017.
Suk, H.-I., Lee, S.-W., Shen, D., and Initiative, A. D. N., othersHierarchical feature representation and multimodal fusion with deep learning for AD/MCI diagnosis. Neuroimage 101:569–582, 2014.
Sarraf, S., Tofighi, G., Deep learning-based pipeline to recognize Alzheimer’s disease using fMRI data, in Future Technologies Conference (FTC), pp. 816–820, (2016).
Monti, M. M., Statistical analysis of fMRI time-series: A critical review of the GLM approach. Front. Hum. Neurosci. 5:28, 2011.
Pernet, C. R., The General Linear Model: Theory and Practicalities in Brain Morphometric Analyses, in Brain Morphometry, Springer, pp. 75–85, (2018).
Oghabian, M. A., Batouli, S. A. H., Norouzian, M., Ziaei, M., and Sikaroodi, H., Using functional magnetic resonance imaging to differentiate between healthy aging subjects, Mild Cognitive Impairment, and Alzheimer’s patients. J. Res. Med. Sci. Off. J. Isfahan Univ. Med. Sci. 15(2):84, 2010.
Marchitelli, R., Collignon, O., and Jovicich, J., Test--retest reproducibility of the intrinsic default mode network: Influence of functional magnetic resonance imaging slice-order acquisition and head-motion correction methods. Brain Connect. 7(2):69–83, 2017.
Goto, M., Abe, O., Miyati, T., Yamasue, H., Gomi, T., and Takeda, T., Head motion and correction methods in resting-state functional MRI. Magn. Reson. Med. Sci. 15(2):178–186, 2016.
Rajagopalan, V., and Pioro, E. P., Disparate voxel based morphometry (VBM) results between SPM and FSL softwares in ALS patients with frontotemporal dementia: which VBM results to consider? BMC Neurol. 15(1):32, 2015.
Carp, J., Park, J., Polk, T. A., and Park, D. C., Age differences in neural distinctiveness revealed by multi-voxel pattern analysis. Neuroimage 56(2):736–743, 2011.
Coutanche, M. N., Thompson-Schill, S. L., and Schultz, R. T., Multi-voxel pattern analysis of fMRI data predicts clinical symptom severity. Neuroimage 57(1):113–123, 2011.
Suk, H.-I., Shen, D., Deep learning-based feature representation for AD/MCI classification, in International Conference on Medical Image Computing and Computer-Assisted Intervention, pp. 583–590, (2013).
Zhu, X., Suk, H.-I., and Shen, D., A novel matrix-similarity based loss function for joint regression and classification in AD diagnosis. Neuroimage 100:91–105, Oct. 2014.
Li, F., Tran, L., Thung, K.-H., Ji, S., Shen, D., and Li, J., A robust deep model for improved classification of AD/MCI patients. IEEE J. Biomed. Heal. Informatics 19(5):1610–1616, Sep. 2015.
S. Liu, S. Liu, W. Cai, H. Che, S. Pujol, R. Kikinis, D. Feng, M. J. Fulham, and others, “Multimodal neuroimaging feature learning for multiclass diagnosis of Alzheimer’s disease,” IEEE Trans. Biomed. Eng., vol. 62, no. 4, pp. 1132–1140, 2015.
Payan, A., Montana, G., Predicting Alzheimer’s disease: a neuroimaging study with 3D convolutional neural networks, arXiv Prepr. arXiv1502.02506, (2015).
Liu, M., Zhang, D., Adeli, E., and Shen, D., Inherent structure-based Multiview learning with multitemplate feature representation for Alzheimer’s disease diagnosis. IEEE Trans. Biomed. Eng. 63(7):1473–1482, Jul. 2016.
Zu, C., Jie, B., Liu, M., Chen, S., Shen, D., Zhang, D., and the A. D. N. Initiative, Label-aligned multi-task feature learning for multimodal classification of Alzheimer’s disease and mild cognitive impairment. Brain Imaging Behav. 10(4):1148–1159, Dec. 2016.
S. Sarraf, G. Tofighi, and for the A. D. N. Initiative, “DeepAD: Alzheimer’s Disease Classification via Deep Convolutional Neural Networks using MRI and fMRI,” bioRxiv, p. 070441, Aug. 2016.
Li, F., Cheng, D., Liu, M., Alzheimer’s disease classification based on combination of multi-model convolutional networks, in Imaging Systems and Techniques (IST), 2017 IEEE International Conference on, pp. 1–5, (2017).
Amoroso, N., Diacono, D., Fanizzi, A., La Rocca, M., Monaco, A., Lombardi, A., Guaragnella, C., Bellotti, R., and Tangaro, S., Deep learning reveals Alzheimer’s disease onset in MCI subjects: Results from an international challenge. J. Neurosci. Methods 302:3–9, 2018.
Liu, M., Cheng, D., Wang, K., Wang, Y.A. D. N., Initiative, and others, “Multi-Modality Cascaded Convolutional Neural Networks for Alzheimer’s Disease Diagnosis,” Neuroinformatics, pp. 1–14, (2018).
C. Yang, A. Rangarajan, and S. Ranka, “Visual Explanations From Deep 3D Convolutional Neural Networks for Alzheimer’s Disease Classification,” arXiv Prepr. arXiv1803.02544, 2018.
S.-H. Wang, P. Phillips, Y. Sui, B. Liu, M. Yang, and H. Cheng, “Classification of Alzheimer’s Disease Based on Eight-Layer Convolutional Neural Network with Leaky Rectified Linear Unit and Max Pooling,” J. Med. Syst., vol. 42, no. 5, p. 85, 2018.
A. Khvostikov, K. Aderghal, J. Benois-Pineau, A. Krylov, and G. Catheline, “3D CNN-based classification using sMRI and MD-DTI images for Alzheimer disease studies,” arXiv Prepr. arXiv1801.05968, 2018.
Shi, J., Zheng, X., Li, Y., Zhang, Q., and Ying, S., Multimodal neuroimaging feature learning with multimodal stacked deep polynomial networks for diagnosis of Alzheimer’s disease. IEEE J. Biomed. Heal. informatics 22(1):173–183, 2018.
S. Liu, S. Liu, W. Cai, S. Pujol, R. Kikinis, and D. Feng, “Early diagnosis of Alzheimer’s disease with deep learning,” in Biomedical Imaging (ISBI), 2014 IEEE 11th International Symposium on, 2014, pp. 1015–1018.
Y. Kazemi and S. Houghten, “A deep learning pipeline to classify different stages of Alzheimer’s disease from fMRI data,” in 2018 IEEE Conference on Computational Intelligence in Bioinformatics and Computational Biology (CIBCB), 2018, pp. 1–8.
“ADNI | Alzheimer’s Disease Neuroimaging Initiative.” .
S. T. Creavin, S. Wisniewski, A. H. Noel-Storr, C. M. Trevelyan, T. Hampton, D. Rayment, V. M. Thom, K. J. E. Nash, H. Elhamoui, R. Milligan, A. S. Patel, D. V Tsivos, T. Wing, E. Phillips, S. M. Kellman, H. L. Shackleton, G. F. Singleton, B. E. Neale, M. E. Watton, et al., “Mini-mental state examination (MMSE) for the detection of dementia in clinically unevaluated people aged 65 and over in community and primary care populations,” Cochrane Database Syst. Rev., Jan. 2016.
Kim, J. W., Byun, M. S., Sohn, B. K., Yi, D., Seo, E. H., Choe, Y. M., Kim, S. G., Choi, H. J., Lee, J. H., Chee, I. S., Woo, J. I., and Lee, D. Y., Clinical dementia rating orientation score as an excellent predictor of the progression to Alzheimer’s disease in mild cognitive impairment. Psychiatry Investig. 14(4):420–426, Jul. 2017.
C. Rorden, “dcm2nii DICOM to NIfTI conversion.” 2012.
Jenkinson, M., Beckmann, C. F., Behrens, T. E. J., Woolrich, M. W., and Smith, S. M., FSL. Neuroimage 62(2):782–790, Aug. 2012.
Woolrich, M. W., Jbabdi, S., Patenaude, B., Chappell, M., Makni, S., Behrens, T., Beckmann, C., Jenkinson, M., and Smith, S. M., Bayesian analysis of neuroimaging data in FSL. Neuroimage 45(1):S173–S186, 2009.
Smith, S. M., Fast robust automated brain extraction. Hum. Brain Mapp. 17(3):143–155, 2002.
Jenkinson, M., Bannister, P., Brady, M., and Smith, S., Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage 17(2):825–841, 2002.
Woolrich, M. W., Ripley, B. D., Brady, M., and Smith, S. M., Temporal autocorrelation in univariate linear modeling of FMRI data. Neuroimage 14(6):1370–1386, 2001.
Jenkinson, M., and Smith, S., A global optimisation method for robust affine registration of brain images. Med. Image Anal. 5(2):143–156, 2001.
Greve, D. N., and Fischl, B., Accurate and robust brain image alignment using boundary-based registration. Neuroimage 48(1):63–72, 2009.
K. He, X. Zhang, S. Ren, and J. Sun, “Deep residual learning for image recognition,” in Proceedings of the IEEE conference on computer vision and pattern recognition, 2016, pp. 770–778.
He, K., Zhang, X., Ren, S., and Sun, J., Identity mappings in deep residual networks. Cham: Springer, 2016, 630–645.
K. Simonyan and A. Zisserman, “Very deep convolutional networks for large-scale image recognition,” arXiv Prepr. arXiv1409.1556, 2014.
C. Szegedy, W. Liu, and Y. Jia, “C. Szegedy, W. Liu, Y. Jia, P. Sermanet, S. Reed, D. Anguelov, D. Erhan, V. Vanhoucke, and A. Rabinovich, arXiv: 1409.4842.”
J. Yosinski, J. Clune, Y. Bengio, and H. Lipson, “How transferable are features in deep neural networks?” pp. 3320–3328, 2014.
A. Sharif Razavian, H. Azizpour, J. Sullivan, and S. Carlsson, “CNN Features Off-the-Shelf: An Astounding Baseline for Recognition.” pp. 806–813, 2014.
Russakovsky, O., Deng, J., Su, H., Krause, J., Satheesh, S., Ma, S., Huang, Z., Karpathy, A., Khosla, A., Bernstein, M., Berg, A. C., and Fei-Fei, L., ImageNet large scale visual recognition challenge. Int. J. Comput. Vis. 115(3):211–252, Dec. 2015.
I. H. Witten, E. Frank, and M. a Hall, Data Mining: Practical Machine Learning Tools and Techniques (Google eBook). 2011.
Acknowledgments
This work was supported by NRPU-4223 from HEC Pakistan. Data collection and sharing for this project was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd. and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California. This work was also supported by Artificial Intelligence and Data Analytics (AIDA) Lab Prince Sultan University Riyadh Saudi Arabia. Authors are thankful for the support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical Approval
We declare that all human and animal studies have been approved by the Medical University of South Carolina Institutional Review Board and have therefore been performed in accordance with the ethical standards laid down in the 1964 Declaration of Helsinki and its later amendments.
Informed Consent
We declare that all patients gave informed consent prior to inclusion in this study.
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Image & Signal Processing
Rights and permissions
About this article
Cite this article
Ramzan, F., Khan, M.U.G., Rehmat, A. et al. A Deep Learning Approach for Automated Diagnosis and Multi-Class Classification of Alzheimer’s Disease Stages Using Resting-State fMRI and Residual Neural Networks. J Med Syst 44, 37 (2020). https://doi.org/10.1007/s10916-019-1475-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10916-019-1475-2