Abstract
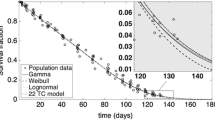
Most mathematical models developed for the survival of haematological cell populations, in particular red blood cells (RBCs), follow the principle of parsimony. They focus on the predominant destruction mechanism of age-related cell death (senescence) and do not account for within subject variability in the RBC lifespan. However, assessment of the underlying physiological destruction mechanisms can be of interest in pathological conditions that affect RBC survival, for example sickle cell anaemia or anaemia of chronic kidney disease. We have previously proposed a semi-mechanistic RBC survival model which accounts for four different types of RBC destruction mechanisms. In this work, it is shown that the proposed model in combination with informative RBC survival data is able to provide a deeper insight into RBC destruction mechanisms. The proposed model was applied in a non-linear mixed effect modelling framework to biotin derived RBC survival data available from literature. Three mechanisms were estimable based on the available data of twelve subjects, including random destruction, senescence and destruction due to delayed failure. It was possible to identify three subjects with a decreased RBC survival in the study population. These three subjects all showed differences in the contribution of the estimated destruction mechanisms: an increased random destruction, versus an accelerated senescence, versus a combination of both.







Similar content being viewed by others
References
Krzyzanski W, Ramakrishnan R, Jusko W (1999) Basic pharmacodynamic models for agents that alter production of natural cells. J Pharmacokinet Biopharm 27(5):467–489
Krzyzanski W, Woo S, Jusko W (2006) Pharmacodynamic models for agents that alter production of natural cells with various distributions of lifespans. J Pharmacokinet Pharmacodyn 33(2):125–166
Freise K, Widness J, Schmidt R, Veng-Pedersen P (2007) Pharmacodynamic analysis of time-variant cellular disposition: reticulocyte disposition changes in phlebotomized sheep. J Pharmacokinet Pharmacodyn 34(4):519–547
Krzyzanski W, Perez-Ruixo J, Vermeulen A (2008) Basic pharmacodynamic models for agents that alter the lifespan distribution of natural cells. J Pharmacokinet Pharmacodyn 35(3):349–377
Lledó-García R, Kalicki R, Uehlinger D, Karlsson M (2012) Modeling of red blood cell life-spans in hematologically normal populations. J Pharmacokinet Pharmacodyn online first: 1–10. doi:10.1007/s10928-012-9261-5
Korell J, Coulter C, Duffull S (2011) A statistical model for red blood cell survival. J Theor Biol 268(1):39–49. doi:10.1016/j.jtbi.2010.10.010
Berlin N, Waldmann T, Weissmann S (1959) Life span of red blood cell. Physiol Rev 39(3):577–616
Korell J, Coulter C, Duffull S (2011) Evaluation of red blood cell labelling methods based on a statistical model for red blood cell survival. J Theor Biol 291:88–98. doi:10.1016/j.jtbi.2011.09.016
Korell J, Vos F, Coulter C, Schollum J, Walker R, Duffull S (2011) Modeling red blood cell survival data. J Pharmacokinet Pharmacodyn 38(6):787–801. doi:10.1007/s10928-011-9220-6
Mock D, Lankford G, Widness J, Burmeister L, Kahn D, Strauss R (1999) Measurement of red cell survival using biotin-labeled red cells: validation against 51Cr-labeled red cells. Transfusion 39(2):156–162
Cohen R, Franco R, Khera P, Smith E, Lindsell C, Ciraolo P, Palascak M, Joiner C (2008) Red cell life span heterogeneity in hematologically normal people is sufficient to alter HbA1c. Blood 112(10):4284–4291
Lavielle M (2005) MONOLIX 1.1 User manual. (Laboratoire de Mathematiques, Universite Paris-Sud, Orsay (France), 2005)
Collett D (2003) Modelling survival data in medical research, 2nd edn. Chapman & Hall/CRC, Boca Raton
Necheles T, Weinstein I, Leroy G (1953) Radioactive sodium chromate for the study of survival of red blood cells: I. The effect of radioactive sodium chromate on red cells. J Lab Clin Med 42(3):358–367
Ebaugh F, Emerson C, Roos J (1953) The use of radioactive chromium 51 as an erythrocyte tagging agent for the determination of red cell survival in vivo. J Clin Invest 29(12):1260–1276
Znojil V (1983) An ignored error in the mathematical formulation of erythrocyte survival curves. J Theor Biol 102(4):625–628. doi:10.1016/0022-5193(83)90395-8
Bentley S, Glass H, Lewis S, Szur L (1974) Elution correction in 51Cr red cell survival studies. Br J Haematol 26(2):179–184
Szymanski I, Valeri C (1970) Factors influencing chromium elution from labelled red cells in vivo and the effect of elution on red-cell survival measurements. Br J Haematol 19(3):397–409
Acknowledgments
J. K. received a University of Otago Prestigious PhD Scholarship and Publishing Bursary.
Author information
Authors and Affiliations
Corresponding author
Appendix
Appendix
Model for random labelling using biotin
Based on Eq. 1 and a constant RBC production rate p on any day τ, the survival of multiple RBC cohorts is given based on Eq. 6. Here, a cohort of RBCs refers to a group of RBCs born on the same day, and N(t) is the total number of RBCs present at day t.
The solution of Eq. 6 was approximated as:
As random loss of the biotin label from viable RBCs is considered to be minimal [10], no adjustment of the proposed RBC survival model to account for flaws associated with the labelling method was required for this study. We have previously described that such an ideal random labelling method can be simulated based on Eq. 6 by stopping the production of RBCs (equivalent to the day of labelling), provided that N(t) has reached steady state before the production is stopped [8]. Observing the disappearance of the cells is then equivalent to the disappearance of the label over time after labelling under the assumption that 1 unit RBCs equals 1 unit label. The constant production rate p can be chosen arbitrarily as only the relative fraction of label left in the circulation is of interest. This relative survival fraction is calculated as N(t)/N(t L ), where t L is the time of labelling.
Rights and permissions
About this article
Cite this article
Korell, J., Duffull, S.B. A semi-mechanistic red blood cell survival model provides some insight into red blood cell destruction mechanisms. J Pharmacokinet Pharmacodyn 40, 469–478 (2013). https://doi.org/10.1007/s10928-013-9322-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10928-013-9322-4