Abstract
Klebsiella pneumoniae and Mycobacterium tuberculosis coinfection is one of the most lethal combinations that has been becoming frequent yet, not diagnosed and reported properly. Due to the simultaneous occurrence of both infections, diagnosis is delayed leading to inadequate treatments and mortality. With the rise of MDR Klebsiella and Mycobacterium, a prophylactic and an immunotherapeutic vaccine has to be entailed for preemptive and adroit therapeutic approach. In this study, we aim to implement reverse vaccinology approach that encompasses a comprehensive evaluation of vital aspects of the pathogens to explore immunogenic epitopes against Omp A of Klebsiella and Rv1698, Rv1973 of Mtb that may help in vaccine development. The designed multi-epitopic vaccine was assessed for antigenicity, allergenicity and various physiochemical parameters. Molecular docking and simulations were executed to assess the immunogenicity and complex stability of the vaccine. The final multi-epitopic vaccine is validated to be highly immunogenic and can serve as a valuable proactive remedy for subject pathogens.
Similar content being viewed by others
Introduction
Coinfection is the concurrent infection of host caused by multiple pathogens. The coinfection of M. tuberculosis and K. pneumoniae has not been frequently accounted. The coincidence of these two infections leads to delayed diagnosis and inadequate treatment causing morbidity and mortality.
Klebsiella pneumoniae is an encapsulated Gram negative organism, commensal to mucosal lining of mammals, human gastrointestinal tract causing nosocomial pneumonia, UTI's, bloodstream infections and sepsis (Pichavant et al. 2018). Klebsiella species are ubiquitous opportunistic pathogens residing in soil and water, having the ability to colonize onto medical devices and in health care settings (Bengoechea and Pessoa 2018). The infections caused by K. pneumonia are common among pediatric wards, elderly and compromised individuals within the healthcare environment (Mcallister-hollod et al. 2014).
Mycobacterium tuberculosis on the other hand is an acid-fast bacillus responsible for tuberculosis (TB) which is the 10th prominent reason of fatality in the world. In 2017, 10 million new cases and 1.6 million deaths were reported due to TB (Vilchèze and Jacobs 2019). According to an estimation by CDC, one fourth of the world’s population is infected with Mtb with children being at primary risk (https://www.cdc.gov/tb/statistics/). TB until now was a curable disease, but with the emergence of multidrug resistant TB, the treatment has become complex that has side effects and can be as long as 2 years (Törün et al. 2005). Unfortunately, Pakistan ranks 5th with highest TB burden and 4th with highest prevalence of MDR TB, amongst the world (Bakuła et al. 2019).
A study conducted in Japan in 2016, reported 128 cases of TB with coexistence of infectious agents. Amongst which the second highest coinfection was with K. pneumoniae that exhibited significant mortality rate than the others (Ishikawa et al. 2019). Coexisting Klebsiella and Mycobacteria infections have been reported in India and worldwide as well (Arora et al. 2015; Attia et al. 2019; Ekwueme et al. 2016; Rafailidis et al. 2008). Coinfection of antibiotic resistant K. pneumoniae and M. tuberculosis is becoming a serious healthcare issue that requires immediate scientific consideration (Arora et al. 2015).
For removal of Klebsiella from human lungs, effective host defense mechanisms are required where the surface of bacteria plays an important role. The three major components of bacterial outer wall which are said to be triggering the immunity include LPS, outer membrane proteins and proteoglycans (Pichavant et al. 2018). Omps are involved in membrane stability, molecular transport and pathogenesis (Choi and Lee 2019; Sousa 2019). Outer membrane protein A (Omp A) which is the member of class of proteins is found to be highly conserved among Enterobacteriaceae (Llobet et al. 2009). Omp A, a well characterized outer membrane protein, has been shown to mediate invasion of eukaryotic cells, facilitate serum resistance and protection of bacteria against host lung defenses (Llobet et al. 2009). Omp A proteins have significant pathogenic roles that include adhesion and invasion of bacteria, intracellular survival as well as evasion of host defenses and stimulators of cytokine production (Confer and Ayalew 2013). The activation of airway epithelial cells via acting on NF-κB, p38 and p44/42-dependent pathways is prevented via K. pneumoniae. Omp A, therefore causing the attenuation of the airway epithelial cell-mediated inflammatory response. Previous studies have shown that detection of K. pneumoniae Omp A by innate immune system aids to clear the infection. Blocking Omp A could not only render bacterial infection but can also help to develop new targets for antimicrobials (Llobet et al. 2009).
Over the years, M. tuberculosis (Mtb) has evolved as fully capable of surviving inside the host cell by evading the immune responses. The cell envelope of Mycobacteria, being an essential virulence factor constitutes of mycolic acids, complex lipids and unique type VII secretion system attributing to complexity of tuberculosis pathogenicity (Madacki et al. 2019; Siroy et al. 2008). The Gram-negative bacteria have various well studied outer membrane proteins that functionalize the outer membrane however, only few outer membrane proteins of Mycobacterium have been reported so far. Rv1698 and Rv1973 were detected to be localized in outer membranes of Mycobacterium that are present in triple the number as compared to the other outer membrane proteins (Boot and Hesper Rego 2019; Song et al. 2008).
The novel and successful bioinformatics approaches have provided us with enormous information on genes of various pathogenic species. With reverse vaccinology approaches; we can use this information for antigen discovery and vaccine development (Del Tordello et al. 2017). Epitopes are considered important to both clinical and biomedical researchers because of their potential for vaccine design against variable and rapidly mutating pathogens (Soria-Guerra et al. 2015).
With the emergence of coinfection of MDR Klebsiella and Mtb and its ensuing mortality, novel and viable vaccine for preemptive protection must be sorted constantly. Since innate immune system targets Omps, we have implemented reverse vaccinology approach that involves a thorough analysis of important aspects of pathogen to explore immunogenic epitopes against Omp A of Klebsiella and RV1698, Rv1973 of Mtb that may help in vaccine development (Boot and Hesper Rego 2019; Rashid et al. 2017).
Materials and Methods
Protein Selection and Sequence Retrieval for Epitope Prediction
Outer membrane proteins were selected for epitope prediction because of their high expression in bacterial cell wall, conservation and active involvement in growth and virulence (Jeannin et al. 2002). Klebsiella pneumoniae is reported to escape the host immune mechanisms by avoiding early detection by circumventing its way around TLRs signaling (Tomás et al. 2015) and subsequently escaping macrophage mediated phagocytosis (Cano et al. 2013). The epitope based vaccines are successfully being pursued by various research groups particularly for targeting conserved epitopes in variable or rapidly mutating pathogens (Soria-Guerra et al. 2015). Omps are under evaluation as potential vaccine candidates (Confer and Ayalew 2013). The FASTA sequence of Omp A, Rv1698 and Rv1973 were obtained from UniprotKB (https://www.uniprot.org). Vaxign (Vaccine Design), a vaccine target prediction and analysis system that employs the principle of reverse vaccinology, was used to check adhesion probability and transmembrane helices of the finalized proteins (Xiang and He 2013).
Epitope Prediction
B Cell Epitope
B lymphocytes have surface receptors that detect B cell epitopes as antigenic elements and differentiate into memory cells and plasma cells. For prediction of B cell epitopes, an online server BCPRED 2.0 was used. Later on, the predicted B cell epitopes were evaluated for antigenicity, surface accessibility, flexibility, beta-turn prediction and hydrophilicity (Chauhan et al. 2019).
CTL Epitope
Using the NetCTL.1.2 server, 9mer length T cell epitopes were predicted, recognizable by commonly occurring HLA I alleles in human population that are likely to encompass > 90% of overall population of the world. The epitopes having consensus score of ≤ 2 were contemplated as strong binders and were further analyzed (Moise et al. 2009).
HTL Epitope
Helper T lymphocyte (HTL) have crucial part in the initiation of both cellular and humoral immune responses (Alexander et al. 1998). Net MHC II pan 3.2 server was used to predict 15-mer length epitopes, identified by HLA II alleles that are likely to cover over 95% of the human population. Peptides with threshold of 2% were categorized as strong binders (Jensen et al. 2018).
Conservation of Selected Proteins and Epitopes
To check the conservation of chosen proteins and predicted epitopes for the vaccine design, a BLASTp analysis was carried out. The analysis was performed to ensure that the chosen proteins are highly conserved in the pathogenic strains only and their predicted epitopes do not share homology with the Homo sapiens.
Selection of Adjuvant and Protein Linker for Vaccine Construction
Adjuvants boost the efficacy of vaccines, enabling small doses of antigen eliciting an effective initial immune response as well as enhanced memory cell differentiation (Khurana et al. 2010). Cholera toxin subunit B (CTB) is considered as a powerful mucosal adjuvant that significantly enhances the immunogenicity of linked exogenous antigens (Klein et al. 2014). CTB is used as a powerful immune activator stimulating innate and acquired immune responses. For linking HTL and CTL epitopes, GPGPG and AAY linkers were respectively utilized (Chauhan et al. 2019). Structure for multi-epitope construct was generated using I-TASSER (Yang and Zhang 2015).
IFN-γ Inducing Epitope Prediction
Interferon gamma (IFN-γ), plays a very important role in adaptive and innate immune responses by stimulation of macrophages and natural killer cells leading to amplified response to MHC antigens. IFN epitope server (http://crdd.osdd.net/raghava/ifnepitope/scan.php) was used for prediction of 15mer IFN-γ epitopes for the designed peptide. Separate epitope predictions were carried for the adjuvant and for the main vaccine peptide owing to the restrictions in number of residues that can be used by the server for prediction. The IFN-γ epitopes are predicted using overlapped sequences assembled by the server using motif and support vector machine (SVM) hybrid approach. This server utilizes a dataset that constitutes of IFN-γ inducing and non-inducing MHC class-II binder that have the ability to activate T-helper cells.
Prediction of Antigenicity and Physicochemical Properties of Vaccine Construct
For each target protein, antigenicity was evaluated by ANTIGENpro and VaxiJen 2.0 server using default parameters to screen probable non-antigenic sequences. On the basis of physicochemical properties, Vaxijen server performed alignment-independent prediction of protective antigens (Dar et al. 2016; Doytchinova and Flower 2008). The ExPASy Protparam tool was used to assess the physiochemical properties of vaccine construct (http://web.expasy.org/protparam). In order to predict the non-allergic behavior of the vaccine construct, AlgPred v.2.0 and AllerTOP v.2.0 servers were utilized (Dimitrov et al. 2014; Saha and Raghava 2006). Proso II Protein-sol software was further used to analyze the solubility of the anticipated vaccine (Hebditch et al. 2017; Smialowski et al. 2012).
Secondary Structure Prediction, 3D Homology Modelling and Refinement of Protein
For the prediction of alpha helices, beta-sheets and coil structure, PSIPRED v3.3 and Raptor X were used (Buchan and Jones 2019; Källberg et al. 2012). I-TASSER was used to model the 3D structure by comparing similarity index between target protein and available template structure provided via PDB (Yang and Zhang 2015). In order to enhance the accurateness of predicted vaccine candidate, refinement was performed via GalaxyRefine (Heo et al. 2013). The refined structure was later on validated by generating Ramachandran plot assessment using RAMPAGE (Lovell et al. 2003) and the quality of the protein model was analyzed through the plotting of z-score via ProSA-Web followed by its 3D validation by using the PROCHECK server (Furey et al. 2006).
Disulfide Engineering to Ensure Protein Stability
Disulfide bonds are the covalent interactions that stabilize tertiary structure of the protein. Disulfide engineering works by creating disulfide bonds into target proteins attributing considerable stability of target protein. Disulfide by Design 2.0 was used to accomplish disulfide engineering on the vaccine construct (Craig and Dombkowski 2013).
Molecular Docking of Vaccine with TLR2 and TLR4 Receptors
TLRs (Toll Like Receptors) are family of transmembrane proteins which are imperative for identification of PAMP (Pathogen Associated Molecular Patterns) expressed by infectious organisms that results in the activation of immune response (Aderem and Ulevitch 2000). Host defense relies on TLR4 and TLR2 during K pneumoniae and M. tuberculosis infection that increases their levels in human airway epithelial cells (Faridgohar and Nikoueinejad 2017; Wieland et al. 2011). The structures for TLR 2 and TLR 4 were obtained from RSCB PDB.
Codon Optimization
For a successful protein expression, various factors are important amongst which codon optimization plays a vital role especially when a heterologous system is used for protein expression. For this purpose, E. coli strain K12 was chosen to host for cloning the vaccine construct. Due to dissimilarity of codon usage between humans and E.coli, a codon adaptation tool (JCAT) was utilized so that the codon usage can be adapted to prokaryotic organisms, thus accelerating the expression rate (Grote et al. 2005).
Molecular Simulation Based on Normal Mode Analysis
For predicting the flexibility of the designed vaccine model, molecular simulation based on the normal mode analysis was carried out which analyzed the protein docked with the TLR2 and TLR4 receptors on the basis of several parameters that included mobility, B-factor, eighen value, covariance and elasticity. The simulation was carried out by employing the iModS server (López-Blanco et al. 2014).
Results
Protein Selection & Sequence Retrieval for Epitope Prediction
Omps are exposed surface porin proteins that are present in high number in the outer membrane of Klebsiella and Mycobacteria (Confer and Ayalew 2013; Siroy et al. 2008). Bacterial outer membrane proteins are characteristically extremely stable and are composed of transmembrane antiparallel β-barrels. They have been shown to have major functional roles that not only include specific and non-specific porin proteins, enzymes, active transporters, structural proteins, but are also implicated in virulence and pathogenesis. Due to their structural simplicity and evident role in pathogenesis, we have investigated Omp A, Rv1698 and Rv1973 as potential multi-epitopic vaccine candidates (Boot and Hesper Rego 2019; Domene, Bond, and Sansom 2003).
Epitope Prediction
Epitope Prediction of B-cell
B cells play primary role in humoral immunity, therefore, an epitope against B cell receptors plays vital role in vaccine production generating antibody production. BCPREDS server predicted 7, 2 and 1 epitopes for Omp A, Rv1698 and Rv1973, respectively. Total 10 B cell epitopes of 20mer length were finalized to be further screened for the vaccine construct.
Epitope Prediction for Cytotoxic T-lymphocytes (CTL)
B and T cells are the first components of immune system to encounter and repel the infectious agent. CTL are CD48 + killer cells that function by killing host target cells with bacterial infection (Maglione and Chan 2009). Using NetCTL 1.2 server, a total of 336, 306 and 152 CTL specific 9mer length epitopes were predicted for Omp A, Rv1698 and Rv1973, respectively. As per standard of IEDB (Calis et al. 2013) the immunogenicity was determined by selecting epitopes with higher scores indicating greater chances to stimulate an immune response. Finally, a total of 10 CTL epitopes for Omp A and 6 each for Rv1698 and Rv1973 respectively, with high immunogenicity scores were utilized for designing the vaccine construct.
Epitope Prediction for Helper T-lymphocyte (HTL)
HTL, the key players of immune system, greatly contribute in orchestrating B cell responses and T cell activation (Maglione and Chan 2009). Thus, the three proteins were subjected to Net MHC II pan 3.2 server for the prediction of HTL receptor specific epitopes. The server predicted 330, 300 and 146 epitopes of 15mer length for Omp A, Rv1698 and Rv1973, respectively. The epitopes with lower percentile valve and IC50 value are the most immunogenic (Ali et al. 2017) that lead to the selection of 7 epitopes for Omp A, 10 epitopes for RV1698 and 4 epitopes for RV1973 for the further screening of the vaccine.
Analysis of the Conservation of Selected Proteins and Epitopes
The BLASTp analysis of the chosen proteins and predicted epitopes against the UNIPROT database indicated that these are highly conserved in the pathogenic strains and share no homology with Homo sapiens (Supplementary Tables 1–2). This is crucial for the safe design of the vaccine so that it can initiate an effective immune response in the human body.
Selection of Adjuvant and Protein Linker for Vaccine Construction
The final vaccine construct was of 350 amino acids that consisted of 03 B cell epitopes, 06 CTL and 03 HTL epitopes with highest immunogenicity values. Each joint has suitable linkers as described previously (Chauhan et al. 2019). The CTB adjuvant was linked to the B-cell epitopes of the three proteins by an EAAAK linker. The intra-B cell epitopes and intra-HTL epitopes were linked via GPGPG linker while intra-CTL epitopes were linked by AAY linker ending with a 6 × His tag (Fig. 1). IFN-γ inducing epitope prediction.
Using the MERCI software, 143 putative IFN-γ inducing epitopes of 15-mer length were predicted for the designed vaccine construct by IFN epitope server. A total of 37 IFN-γ inducing epitopes having positive scores for the main vaccine sequence were predicted.
Prediction of Antigenicity and Physicochemical Properties of Vaccine Construct
The predicted probability of antigenicity by ANTIGENpro is 0.874312 while VaxiJen showed it to be 0.6359 with a bacterial model. The ExPASy Protparam tool analyzed the physiochemical properties on the basis of seven parameters (Anon n.d.). The molecular weight was evaluated to be 36 kDa, as reported previously, lower molecular weight will promote the vaccine antigenicity (Khatoon et al. 2017). The theoretical pI was calculated to be 5.78 that showed the slight acidic nature whilst the number of positive and negative charged residues was 24 and 31, respectively. The estimated half-life in mammalian reticulocytes was 30 h, in vitro; while > 20 and > 10 h in yeast and E. coli, in vivo. The instability index was found to be 25.89 classifying the protein as stable. The designed vaccine construct had an aliphatic index of 84.11, the higher the aliphatic index, the higher is the thermostability. The Grand average of hydropathicity (GRAVY) was estimated to be -0.028 that shows that the vaccine construct is hydrophilic in nature (Anon n.d.). AlgPred v.2.0 and AllerTOP v.2.0 servers found that the vaccine is non-allergenic and safe for human use. Protein-sol software further analyzed the solubility to be 0.483 indicating it to be highly soluble in nature. This concludes that the designed vaccine construct is not only immunogenic but also hydrophilic and thermostable by nature.
Secondary Structure Prediction, 3D Homology Modelling and Refinement of Protein
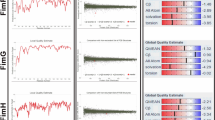
RaptorX and PSIPRED predicted the secondary structures of the vaccine according to which it constituted of 31% alpha helix, 13% beta sheets and 55% random coil (Fig. 2). GalaxyRefine, the protein refinement tool was used to increase the number of residues in favored regions. The refinement (Fig. 3a) was later on validated by Ramachandran plot that presented 90.5% residues in favored region, 70.2% in allowed region and 2.3% in the outlier region (Fig. 3b). The z-score was found to be -2.59 representing good quality of vaccine (Fig. 3c). The 3D validation of the vaccine model was done by PROCHECK server that showed the quality to be 67%.
Disulfide Engineering to Ensure Protein Stability
For the stability of the vaccine, Disulfide by design v2.0 showed 37 residues can be used for disulfide engineering. Only three were selected to mutate the vaccine construct because value of energy should be less than 2.2 and Chi3 values should be between -87 and + 97 (47). Therefore, a total of six mutations were created at the Leu174-Pro176 and Thr183-Asn181 and at Ala321-His345 residue pairs as depicted in Fig. 4.
Molecular Docking of Vaccine with TLR2 and TLR4
TLR plays major role in onset of inflammation response by recognition of the pathogen. Host defense against K. pneumoniae and M. tuberculosis infection greatly relies on TLR2 and TLR4 (Regueiro et al. 2009; Faridgohar and Nikoueinejad 2017). HADDOCK 2.2 was used for analyzing the favorable interactions between the two molecules. Based on the docking results, favorable interactions were observed between the designed vaccine with the TLR2 and TLR4 receptors. A total number of 14 hydrogen bonds were formed between the vaccine and TLR2 receptor whereas 6 were found between the vaccine and TLR4 immune receptor (Fig. 5).
Optimization of the Codon
Optimization of the codon is essential for expressing the vaccine in an appropriate host model. The designed vaccine sequence was optimized, and the resulting improved sequence was reported to have a GC content of 56% and the Codon Adaptation Index was calculated to be 0.9551. Normally, the GC content should be found between 30–70% whereas the CAI should be close to the ideal value of 1. The predicted values for the vaccine sequence are suitable as they lie within the ideal ranges (Fig. 6).
Molecular Simulation Based on Normal Mode Analysis
For confirming the flexibility of the two complexes i-e. Vaccine docked with TLR2 and Vaccine docked with TLR4, normal mode analysis was carried out which evaluated the subject complexes on the basis of the internal coordinates that depicted the mobility of these protein complexes. Based on the simulation results (Figs. 7, 8), the vaccine complexes were also found to be flexible as their B factor values, Eighen values, co-variances and elastic models showed good results.
Discussion
Tubercular and bacterial coinfection is common in immunocompromised patients with organisms involved in nosocomial infections (Arora et al. 2015). The coexistent infections of Klebsiella and Mycobacteria have been diagnosed but not frequently reported (Feldman 2000). Limited published studies depict the problem of co-infection of tuberculosis with bacterial pneumoniae in regions where there is high prevalence of TB and the treatment is based on presumptive diagnosis (Arora et al. 2015; Feldman 2000; Iliyasu et al. 2018). These infections are difficult to treat since most clinical isolates show resistance to number of available antibiotics that lead to treatment failures and increased risk of systemic dissemination (Jeannin et al. 2002). With emergence of multidrug resistant strains, the effective control of Klebsiella and Mycobacteria infections need immuno-prophylaxis as well as immunotherapy.
Utilization of bioinformatics tools has steered the direction of vaccines towards the employment of isolated epitopes that are able to stimulate concentrated immune responses. Epitopes have been predicted and employed in innovative formulations of vaccine, therefore providing a better alternative to the whole pathogen vaccines (Soria-Guerra et al. 2015). Based on the ability to effectively bind with the both MHCI and MHCII, peptide based vaccines are considered more effective in terms of the epitope predictions (Naz et al. 2015).
Omps perform in various capacities that include maintaining structure and stability of the membrane, signal transduction, soluble transport, catalysis and defense (Jeannin et al. 2002). The study conducted in Japan at the Kyushu University, was the first to integrate the proteins of the outer membrane in formulations of vaccines (Serushago et al. 1989). Up till now, no such vaccine has been reported for coexistent Klebsiella pneumoniae and Mycobacterium tuberculosis. Therefore, keeping these points in consideration, the present study was aimed to design a multi-epitope vaccine for coinfection of Klebsiella and Mycobacteria by targeting the outer membrane proteins Omp A, Rv1698 and Rv1973, which facilitate the bacterial entry and pathogenesis in the host.
Using in silico biological tools, the most critical step is the selection of suitable antigenic epitopes for the target proteins to design a multi-epitope vaccine construct (Chauhan et al. 2019). The selected epitopes (B cell, CTL and HTL) for the target protein in this study have passed several immune filters that have been designed to screen the epitopes. These tests such as antigenicity check, BLASTp analysis, Transmembrane helices prediction and molecular weight prediction ensure that selected epitopes are immunogenic, non-allergenic and promiscuous and they don’t overlap with human proteins. Furthermore, antigenicity and allergenicity of the final vaccine construct was also predicted to ensure its safety. The stability of the vaccine was ensured by Disulphide engineering. After cautious evaluation of energy and Chi3 values, three pairs of amino acid residues were selected for which six mutations were created.
Multi-epitope vaccines are different in concepts having properties as compared to conventional or single-epitope vaccines: they have multiple MHC-restricted epitopes which can be recognized by TLRs and the presence of T and B-cell epitopes can induce both humoral and cellular immune responses (Zhang 2018). TLR2 and TLR4 dependent mediated signals play a critical role in host defense against Klebsiella and Mycobacteria (Faridgohar and Nikoueinejad 2017; Sánchez et al. 2010; Wieland et al. 2011). TLR2 plays a significant role in immune modulation as it helps to locate pathogen associated molecular patterns derived from the microbial cells. Similarly, TLR4 is also involved in host microbial cell interactions in the immune system (Regueiro et al. 2009). Therefore, molecular docking analysis was performed for vaccine construct with TLR2 and TLR4 that analyzed good affinity and favorable interaction with the immune receptors. To evaluate the flexibility and interaction of the vaccine construct normal mode analysis was performed that showed the vaccine to be flexible enough to effortlessly interact with the host environment.
Conclusion
The in-silico approach is the first step towards designing a vaccine for which effective selection of the target protein plays an integral role. In order to improve our knowledge regarding host-microbe interactions and gain vision of their mechanisms, in-silico analysis has enabled us to develop a potential new vaccine candidate. On the basis of these outcomes, this multi-epitope vaccine design can confer protection against Klebsiella and Mycobacteria coinfection with greater specificity and safety.
References
Aderem A, Ulevitch RJ (2000) Toll-like receptors in the induction of the innate immune response. Nature 406(6797):782–787
Alexander J, Fikes J, Hoffman S, Franke E, Sacci J, Appella E, Chisari FV, Guidotti LG, Chesnut RW, Livingston B, Sette A (1998) The optimization of helper T lymphocyte (HTL) function in vaccine development. Immunol Res 18(2):79–92
Ali M, Pandey RK, Khatoon N, Narula A, Mishra A, Prajapati VK (2017) Exploring dengue genome to construct a multi-epitope based subunit vaccine by utilizing immunoinformatics approach to battle against dengue infection. Sci Rep 7(1):9232
Arora AA, Krishnaswamy UM, Moideen RP, Padmaja MS (2015) Tubercular and bacterial coinfection: a case series. Lung India 32(2):172–174
Attia EF, Pho Y, Nhem S, Sok C, By B, Phann D, Nob H, Thann S, Yin S, Noce R, Kim C, Letchford J, Fassier T, Chan S, Eoin West T (2019) Tuberculosis and other bacterial co-infection in cambodia: a single center retrospective cross-sectional study. BMC Pulm Med 19(1):60
Bakuła Z, Javed H, Pleń M, Jamil N, Tahir Z, Jagielski T (2019) Genetic diversity of multidrug-resistant mycobacterium tuberculosis isolates in Punjab, Pakistan. Infect Genet Evol 72:16–24
Bengoechea A, Pessoa JS (2018) Klebsiella pneumoniae infection biology: living to counteract host defences. FEMS Microbiol Rev. https://doi.org/10.1093/femsre/fuy043
Boot M, Hesper Rego EH (2019) Drug susceptibility of individual mycobacterial cells. In: Lewis K (ed) Persister cells and infectious disease. Springer, Berlin, pp 247–272
Buchan DWA, Jones DT (2019) The PSIPRED protein analysis workbench: 20 years on. Nucleic Acids Res 47(W1):W402–W407
Calis JJA, Maybeno M, Greenbaum JA, Weiskopf D, De Silva AD, Sette A, Keşmir C, Peters B (2013) Properties of MHC class I presented peptides that enhance immunogenicity. PLoS Comput Biol 9(10):e1003266
Cano V, Garmendia J, Llobet E, Suárez T, Tomás JM, Moranta D, Bengoechea JA, March C, Pérez-Gutiérrez C (2013) Role of bacterial surface structures on the interaction of Klebsiella pneumoniae with phagocytes. PLoS ONE 8(2):e56847
Chauhan V, Rungta T, Goyal K, Singh MP (2019) Designing a multi-epitope based vaccine to combat kaposi sarcoma utilizing immunoinformatics approach. Sci Rep 9(1):2517
Choi U, Lee C-R (2019) Distinct roles of outer membrane porins in antibiotic resistance and membrane integrity in Escherichia coli. Front Microbiol 10:953
Confer AW, Ayalew S (2013) The OmpA family of proteins: roles in bacterial pathogenesis and immunity. Vet Microbiol 163(3–4):207–222
Craig DB, Dombkowski AA (2013) Disulfide by design 20: a web-based tool for disulfide engineering in proteins. BMC Bioinform 14(1):346
Dar H, Zaheer T, Rehman MT, Ali A, Javed A, Khan GA, Babar MM, Waheed Y (2016) Prediction of promiscuous T-cell epitopes in the Zika virus polyprotein: an in silico approach. Asian Pac J Trop Med 9(9):844–850
Del Tordello E, Rappuoli R, Delany I (2017) Reverse vaccinology: exploiting genomes for vaccine design. Hum Vaccin 65:86
Dimitrov I, Bangov I, Flower DR, Doytchinova I (2014) AllerTOP vol 2—a server for in silico prediction of allergens. J Mol Model 20(6):2278
Domene C, Bond PJ, Sansom MSP (2003) Membrane protein simulations: ion channels and bacterial outer membrane proteins. Adv Protein Chem 66:159–193
Doytchinova IA, Flower DR (2008) Bioinformatic approach for identifying parasite and fungal candidate subunit vaccines. Open Vaccine J 1:4
Ekwueme C, Otu AA, Chinenye S, Unachukwu C, Oputa RN, Korubo I, Enang OE (2016) Haemoptysis in a female with diabetes mellitus: a unique presentation of chronic pulmonary aspergillosis, pulmonary tuberculosis, and Klebsiella peumoniae co-infection. Clin Case Rep 4(4):432–436
Faridgohar M, Nikoueinejad H (2017) New findings of toll-like receptors involved in mycobacterium tuberculosis infection. Pathog Glob Health 111(5):256–264
Feldman C (2000) Klebsiella pneumoniae: pneumonia. Clin Pulm Med 7(6):295–303
Furey W, Cowtan KD, Zhang KYJ, Main P, Brunger AT, Adams PD, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Pannu NS, Read RJ, Rice LM, Simonson T, Tronrud DE, Ten Eyck LF, Lamzin VS, Perrakis A, Wilson KS, Laskowski RA, MacArthur MW, Thornton JM, Kraulis PJ, Richardson DC, Richardson JS, Kabsch W, Sheldrick GM (2006) Programs and program systems in wide use. In: International tables for crystallography. International Union of Crystallography, pp 695–743.
Grote A, Hiller K, Scheer M, Münch R, Nörtemann B, Hempel DC, Jahn D (2005) JCat: a novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res 33(2):W526–W531
Hebditch M, Alejandro Carballo-Amador M, Charonis S, Curtis R, Warwicker J (2017) Protein-sol: a web tool for predicting protein solubility from sequence. Bioinformatics 33(19):3098–3100
Heo L, Park H, Seok C (2013) GalaxyRefine: protein structure refinement driven by side-chain repacking. Nucleic Acids Res 41:W384–W388
Iliyasu G, Mohammad AB, Yakasai AM, Dayyab FM, Oduh J, Habib AG (2018) Gram-negative bacilli are a major cause of secondary pneumonia in patients with pulmonary tuberculosis: evidence from a cross-sectional study in a tertiary hospital in Nigeria. Trans R Soc Trop Med Hyg 112(5):252–254
Ishikawa S, Igari H, Yamagishi K, Takayanagi S, Yamagishi F (2019) Microorganisms isolated at admission and treatment outcome in sputum smear-positive pulmonary tuberculosis. J Infect Chemother 25(1):45–49
Jeannin P, Magistrelli G, Goetsch L, Haeuw JF, Thieblemont N, Bonnefoy JY, Delneste Y (2002) Outer membrane protein a (ompa): a new pathogen-associated molecular pattern that interacts with antigen presenting cells—impact on vaccine strategies. Vaccine 20(Suppl 4):23–27
Jensen KK, Andreatta M, Marcatili P, Buus S, Greenbaum JA, Yan Z, Sette A, Peters B, Nielsen M (2018) Improved methods for predicting peptide binding affinity to mhc class II molecules. Immunology 154(3):394–406
Källberg M, Wang H, Wang S, Peng J, Wang Z, Hui Lu, Jinbo Xu (2012) Template-based protein structure modeling using the RaptorX web server. Nat Protoc 7(8):1511–1522
Khatoon N, Pandey RK, Prajapati VK (2017) Exploring leishmania secretory proteins to design B and T cell multi-epitope subunit vaccine using immunoinformatics approach. Sci Rep 7(1):1–12
Khurana S, Chearwae W, Castellino F, Manischewitz J, King LR, Honorkiewicz A, Rock MT, Edwards KM, Del Giudice G, Rappuoli R, Golding H (2010) Vaccines with MF59 adjuvant expand the antibody repertoire to target protective sites of pandemic Avian H5N1 influenza virus. Sci Transl Med 2(15):15ra5
Klein JS, Jiang S, Galimidi RP, Keeffe JR, Bjorkman PJ (2014) Design and characterization of structured protein linkers with differing flexibilities. Protein Eng Des Select PEDS 27(10):325–330
Llobet E, March C, Giménez P, Bengoechea JA (2009) Klebsiella pneumoniae OmpA confers resistance to antimicrobial peptides. Antimicrob Agents Chemother 53(1):298–302
López-Blanco JR, Aliaga JI, Quintana-Ortí ES, Chacón P (2014) IMODS: internal coordinates normal mode analysis server. Nucleic Acids Res 42(W1):W271–W276
Lovell SC, Davis IW, Arendall WB, De-Bakker PIW, Word JM, Prisant MG, Richardson JS, Richardson DC (2003) Structure validation by Cα geometry: φ, ψ and Cβ deviation. Proteins Struct Funct Genet 50(3):437–450
Madacki J, Fiol GM, Brosch R (2019) “Update on the virulence factors of the obligate pathogen Mycobacterium tuberculosis and related tuberculosis-causing Mycobacteria. Infect Genet Evol 72:67–77
Maglione PJ, Chan J (2009) How B cells shape the immune response against Mycobacterium tuberculosis. Eur J Immunol 39(3):676–686
Mcallister-hollod L, Nadle J, Ray SM, Thompson DL, Wilson LE (2014) Multistate point-prevalence survey of health care-associated infections. N Engl J Med 370(13):1198–1208
Moise L, McMurry JA, Buus S, Frey S, Martin WD, De Groot AS (2009) In silico-accelerated identification of conserved and immunogenic variola/vaccinia T-cell epitopes. Vaccine 27(46):6471–6479
Naz A, Awan FM, Obaid A, Muhammad SA, Paracha RZ, Ahmad J, Ali A (2015) Identification of putative vaccine candidates against helicobacter pylori exploiting exoproteome and secretome: a reverse vaccinology based approach. Infect Genet Evol 32:280–291
Pichavant M, Delneste Y, Jeannin P, Brichet A, Tonnel A-B (2018) Outer membrane protein a from klebsiella pneumoniae activates bronchial epithelial cells: implication in neutrophil recruitment. J Immunol 171(12):6697–6705
Rafailidis PI, Kapaskelis A, Christodoulou C, Galani E, Falagas ME (2008) Concurrent M. tuberculosis, Klebsiella pneumoniae, and Candida albicans infection in liver metastasis of bowel carcinoma. Eur J Clin Microbiol Infect Dis 27(8):753–755
Rashid MI, Naz A, Ali A, Andleeb S (2017) Prediction of vaccine candidates against Pseudomonas aeruginosa: an integrated genomics and proteomics approach. Genomics 109(3–4):274–283
Regueiro V, Moranta D, Campos MA, Margareto J, Garmendia J, Bengoechea JA (2009) Klebsiella pneumoniae increases the levels of toll-like receptors 2 and 4 in human airway epithelial cells. Infect Immun 77(2):714–724
Saha S, Raghava GPS (2006) AlgPred: prediction of allergenic proteins and mapping of IgE epitopes. Nucleic Acids Res 34:W202–W209
Sánchez D, Rojas M, Hernández I, Radzioch D, García LF, Barrera LF (2010) Role of TLR2- and TLR4-mediated signaling in Mycobacterium tuberculosis-induced macrophage death. Cell Immunol 260(2):128–136
Serushago BA, Mitsuyama M, Handa T, Koga T, Nomoto K (1989) Role of antibodies against outer-membrane proteins in murine resistance to infection with encapsulated Klebsiella pneumoniae. Microbiology 135(8):2259–2268
Siroy A, Mailaender C, Harder D, Koerber S, Wolschendorf F, Danilchanka O, Wang Y, Heinz C, Niederweis M (2008) Rv1698 of Mycobacterium tuberculosis represents a new class of channel-forming outer membrane proteins. J Biol Chem 283(26):17827–17837
Smialowski P, Doose G, Torkler P, Kaufmann S, Frishman D (2012) PROSO II—a new method for protein solubility prediction. FEBS J 279(12):2192–2200
Song H, Sandie R, Wang Y, Andrade-Navarro MA, Niederweis M (2008) Identification of outer membrane proteins of Mycobacterium tuberculosis. Tuberculosis 88(6):526–544
Soria-Guerra RE, Nieto-Gomez R, Govea-Alonso DO, Rosales-Mendoza S (2015) An overview of bioinformatics tools for epitope prediction: implications on vaccine development. J Biomed Inform 53:405–414
Sousa MC (2019) New antibiotics target the outer membrane of bacteria. Nature 576(7787):389–390
Tomás A, Lery L, Regueiro V, Pérez-Gutiérrez C, Martínez V, Moranta D, Llobet E, González-Nicolau M, Insua JL, Tomas JM, Sansonetti PJ, Tournebize R, Bengoechea JA (2015) Functional genomic screen identifies klebsiella pneumoniae factors implicated in blocking nuclear factor ΚB (NF-ΚB) signaling. J Biol Chem 290(27):16678–16697
Törün T, Güngor G, Özmen J, Bölükbaşi Y, Maden E, Biçakçi B, Ataç G, Sevim T, Tahaoǧlu K (2005) Side effects associated with the treatment of multidrug-resistant tuberculosis. Int J Tuberc Lung Dis 9(12):1373–1377
Vilchèze C, Jacobs WR (2019) The isoniazid paradigm of killing, resistance, and persistence in Mycobacterium tuberculosis. J Mol Biol 431(18):3450–3461
Wieland CW, van Lieshout MHP, Hoogendijk AJ, van der Poll T (2011) Host defence during klebsiella pneumonia relies on haematopoietic-expressed toll-like receptors 4 and 2. Eur Respir J 37(4):848–857
Xiang Z, He Y (2013) Genome-wide prediction of vaccine targets for human herpes simplex viruses using vaxign reverse vaccinology. BMC Bioinform 14(SUPPL4):S2
Yang J, Zhang Y (2015) I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res 43(W1):W174–W181
Zhang L (2018) Multi-epitope vaccines: a promising strategy against tumors and viral infections. Cell Mol Immunol 15(2):182–184
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Rahmat Ullah, S., Majid, M., Rashid, M.I. et al. Immunoinformatics Driven Prediction of Multiepitopic Vaccine Against Klebsiella pneumoniae and Mycobacterium tuberculosis Coinfection and Its Validation via In Silico Expression. Int J Pept Res Ther 27, 987–999 (2021). https://doi.org/10.1007/s10989-020-10144-1
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10989-020-10144-1