Abstract
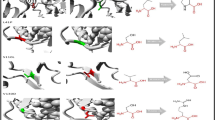
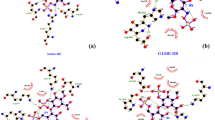
Charcot-Marie-Tooth disease (CMT) is one of the most commonly inherited congenital neurological disorders, affecting approximately 1 in 2500 in the US. About 80 genes were found to be in association with CMT. The phosphoribosyl pyrophosphate synthetase 1 (PRPS1) is an essential enzyme in the primary stage of de novo and salvage nucleotide synthesis. The mutations in the PRPS1 gene leads to X-linked Charcot-Marie-Tooth neuropathy type 5 (CMTX5), PRS super activity, Arts syndrome, X-linked deafness-1, breast cancer, and colorectal cancer. In the present study, we obtained 20 missense mutations from UniProt and dbSNP databases and applied series of comprehensive in silico prediction methods to assess the degree of pathogenicity and stability. In silico tools predicted four missense mutations (D52H, M115 T, L152P, and D203H) to be potential disease causing mutations. We further subjected the four mutations along with native protein to 50 ns molecular dynamics simulation (MDS) using Gromacs package. The resulting trajectory files were analyzed to understand the stability differences caused by the mutations. We used the Root Mean Square Deviation (RMSD), Radius of Gyration (Rg), solvent accessibility surface area (SASA), Covariance matrix, Principal Component Analysis (PCA), Free Energy Landscape (FEL), and secondary structure analysis to assess the structural changes in the protein upon mutation. Our study suggests that the four mutations might affect the PRPS1 protein function and stability of the structure. The proposed study may serve as a platform for drug repositioning and personalized medicine for diseases that are caused by the PRPS1 deficiency.







Similar content being viewed by others
References
Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR (2010) A method and server for predicting damaging missense mutations. Nat Methods 7:248–249
Agrahari A, George Priya Doss C (2015) Impact of I30T and I30M substitution in MPZ gene associated with Dejerine–Sottas syndrome type B (DSSB): a molecular modeling and dynamics. J Theor Biol 382:23–33
Ali SK, Sneha P, Priyadharshini Christy J, Zayed H, George Priya Doss C (2016) Molecular dynamics-based analyses of the structural instability and secondary structure of the fibrinogen gamma chain protein with the D356V mutation. J Biomol Struct Dyn 35(12):2714–2724
Alm E, Baker D (1999) Prediction of protein-folding mechanisms from free-energy landscapes derived from native structures. Proc Natl Acad Sci U S A 96:11305–11310
Amadei A, Linssen AB, Berendsen HJ (1993) Essential dynamics of proteins. Proteins 17:412–425
Arts WF, Loonen MC, Sengers RC, Slooff JL (1933) X-linked ataxia, weakness, deafness, and loss of vision in early childhood with a fatal course. Ann Neurol 33:535–539
Bairoch A, Apweiler R (1996) The SWISS-PROT protein sequence data bank and its new supplement TREMBL. Nucleic Acids Res 24(1):21–25
Bakhit YH, Ibrahim MO, Amin M, Mirghani YA, Hassan MA (2016) In silico analysis of SNPs in PARK2 and PINK1 genes that potentially cause autosomal recessive Parkinson disease. Adv Bioinforma 2016:9313746
Becker MA (2008) Hyperuricemia and Gout In The Online Metabolic and Molecular Bases of Inherited Disease (OMMBID), D Valle, ed New York: McGraw-Hill, chapter 205
Bhardwaj A, Dhar YV, Asif MH, Bag SK (2016) In Silico identification of SNP diversity in cultivated and wild tomato species: insight from molecular simulations. Sci Rep 6:38715
Bian Y, Zhang J, Wang J, Wang J, Wang W, Jiao X (2015) Free energy landscape and multiple folding pathways of an H-type RNA pseudoknot. PLoS One 10:e0129089. https://doi.org/10.1371/journal.pone.0129089
Bromberg Y, Rost B (2007) SNAP: predict the effect of non-synonymous polymorphisms on function. Nucleic Acids Res 35(11):3823–3835
de Brouwer AP, Williams KL, Duley JA, van Kuilenburg AB, Nabuurs SB, Egmont-Petersen M, Lugtenberg D, Zoetekouw L, Banning MJ, Roeffen M (2007) Arts syndrome is caused by loss-of-function mutations in PRPS1. Am J Hum Genet 81:507–518. https://doi.org/10.1007/s11011-017-0006-4.
Capriotti E, Fariselli P, Rossi I, Casadio R (2008) A three-state prediction of single point mutations on protein stability changes. BMC Bioinformatics 9:S6
Da Silva Figueiredo Celestino Gomes P, Chauvot De Beauchêne I, Panel N, Lopez S, De Sepulveda P, Geraldo Pascutti P, Solary E, Tchertanov L (2016) Insight on mutation-induced resistance from molecular dynamics simulations of the native and mutated CSF-1R and KIT. PLoS One 11(7):e0160165
Doss CG, Sethumadhavan R (2009) Investigation on the role of nsSNPs in HNPCC genes--a bioinformatics approach. J Biomed Sci 16:42. https://doi.org/10.1186/1423-0127-16-42
Doss CGP, Chakraborty C, Chen L, Zhu H (2014) Integrating in silico prediction methods, molecular docking, and molecular dynamics simulation to predict the impact of ALK missense mutations in structural perspective. Biomed Res Int 2014:895831. https://doi.org/10.1155/2014/895831
Doss CGP, Alasmar DR, Bux RI, Sneha P, Bakhsh FD, Al-Azwani I, El BR, Zayed H (2016) Genetic epidemiology of Glucose-6-phosphate dehydrogenase deficiency in the Arab world. Sci Rep 6:37284. https://doi.org/10.1038/srep37284
Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723
Hess B, Kutzner C, van der Spoel D, Lindahl E (2004) GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput 4:435–447
Jorgensen WL, Maxwell DS, Tirado-Rives J (1996) Development and testing of the OPLS all-atom force field on conformational energetic and properties of organic liquids. J Am Chem Soc 118:11225–11236
Kabsch W, Sander C (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22:2577–2637
Kim JW, Kim HJ (2012) Charcot-Marie-tooth neuropathy X type 5. Gene Reviews http://wwwncbinlmnihgov/books/NBK1876/
Kim HJ, Sohn KM, Shy ME, Krajewski KM, Hwang M, Park JH, Jang SY, Won HH, Choi BO, Hong SH (2007) Mutations in PRPS1, which encodes the phosphoribosyl pyrophosphate synthetase enzyme critical for nucleotide biosynthesis, cause hereditary peripheral neuropathy with hearing loss and optic neuropathy (cmtx5). Am J Hum Genet 81:552–558
Kumar P, Henikoff S, Ng PC (2009) Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc 4:1073–1081
Li S, Lu Y, Peng B, Ding J (2007) Crystal structure of human phosphoribosylpyrophosphate synthetase 1 reveals a novel allosteric site. Biochem J 401(1):39–47
Lobanov MI, Bogatyreva NS, Galzitskaia OV (2008) Radius of gyration is indicator of compactness of protein structure. Mol Biol (Mosk) 42:701–706
McMurry J (2012) Organic chemistry. Brooks/Cole, Cengage Learning
Minde DP, Anvarian Z, Rüdiger SG, Maurice MM (2011) Messing up disorder: how do missense mutations in the tumor suppressor protein APC lead to cancer? Mol Cancer 10:101. https://doi.org/10.1186/1476-4598-10-101
Mittal R, Patel K, Mittal J, Chan B, Yan D, Grati M, Liu XZ (2015) Association of PRPS1 mutations with disease phenotypes. Dis Markers 2015:127013. https://doi.org/10.1155/2015/127013
Mosaeilhy A, Mohamed MM, C GPD et al (2017) Genotype-phenotype correlation in 18 Egyptian patients with glutaric acidemia type I. Metab Brain Dis 32:1417–1426. https://doi.org/10.1007/s11011-017-0006-4
Nagasundaram N, Hailong Z, Jiming L, Karthick V, George Priya Doss C, Chiranjib C, Luonan C (2015) Analysing the effect of mutation on protein function and discovering potential inhibitors of CDK4: molecular modelling and dynamics studies. PLoS One 10(8):e0133969
Ndagi U, Mhlongo NN, Soliman ME (2017) The impact of Thr91 mutation on c-Src resistance to UM-164: molecular dynamics study revealed a new opportunity for drug design. Mol BioSyst 13(6):1157–1171
Rajasekaran R, Sudandiradoss C, Doss CG, Sethumadhavan R (2007) Identification and in silico analysis of functional SNPs of the BRCA1 gene. Genomics 90(4):447–452
Roessler BJ, Bell G, Heidler S, Seino S, Becker M, Palella TD (1990) Cloning of two distinct copies of human phosphoribosylpyrophosphate synthetase cDNA. Nucleic Acids Res 18:193
Sang P, Hu W, Ye YJ, Li LH, Zhang C, Xie YH, Meng ZH (2016) In silico screening, molecular docking, and molecular dynamics studies of SNP-derived human P5CR mutants. J Biomol Struct Dyn 27:1–13
Schreiber G, Buckle AM, Fersht AR, Bycroft M, Fersht AR, Tratschin JG (1994) Stability and function: two constraints in the evolution of barstar and other proteins. Structure (London, England : 1993) 2(10):945–951
Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K (2001) dbSNP: the NCBI database of genetic variation. Nucleic Acids Res29:308–311
Shoichet BK, Baase WA, Kuroki R, Matthews BW (1995) A relationship between protein stability and protein function. Proc Natl Acad Sci 92(2):452–456
Sjöblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, Szabo S, Buckhaults P, Farrell C, Meeh P, Markowitz SD, Willis J, Dawson D, Willson JK, Gazdar AF, Hartigan J, Wu L, Liu C, Parmigiani G, Park BH, Bachman KE, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE (2006) The consensus coding sequences of human breast and colorectal cancers. Science 314:268–274
Sneha P, Doss, George Priya C (2016) Chapter seven – molecular dynamics: new frontier in personalized medicine, in: Advances in protein chemistry and structural biology. 102:181–224. https://doi.org/10.1016/bs.apcsb.2015.09.004
Sneha P, Doss CG (2017) Elucidating the mutational landscape in hepatocyte nuclear factor 1β (HNF1B) by computational approach. Adv Protein Chem Struct Biol 107:283–306
Sneha P, Thirumal Kumar D, Tanwar H, Siva R, George Priya Doss C, Zayed H (2017a) Structural analysis of G1691S variant in the human Filamin B gene responsible for Larsen syndrome: a comparative computational approach. J Cell Biochem 18:1900–1910
Sneha P, Thirumal Kumar D, George Priya C, Doss C, Siva R, Zayed H (2017b) Determining the role of missense mutations in the POU domain of HNF1A that reduce the DNA-binding affinity: a computational approach. PLoS One 12(4):e0174953
Sperling O, Boer P, Persky-Brosh S, Kanarek E, DeVries A (1972) Altered kinetic property of erythrocyte phosphoribosylpsyrophosphate synthetase in excessive purine production. Rev Eur Etud Clin Biol 17:703–706
Stanger HE, Syud FA, Espinosa JF, Giriat I, Muir T, Gellman HS (2000) Length-dependent stability and strand length limits in antiparallel β-sheet secondary structure. Proc Natl Acad Sci U S A 98(21):12015–12020
Taira M, Ishijima S, Kita K, Yamada K, Iizasa T, Tatibana M (1987) Nucleotide and deduced amino acid sequences of two distinct cDNAs for rat phosphoribosylpyrophosphate synthetase. J Biol Chem 262:14867–14870
Tavtigian SV, Deffenbaugh AM, Yin L, Judkins T, Scholl T, Samollow PB, de Silva D, Zharkikh A, Thomas A (2006) Comprehensive statistical study of 452 BRCA1 missense substitutions with classification of eight recurrent substitutions as neutral. J Med Genet 43:295–305
Tekcan A (2016) In silico analysis of FMR1 gene missense SNPs. Cell Biochem Biophys 74:109–127
Teng S, Srivastava AK, Wang L (2010) Sequence feature-based prediction of protein stability changes upon amino acid substitutions. BMC Genomics 11:2–5
Theobald DL, Wuttke DS (2008) Accurate structural correlations from maximum likelihood superpositions. PLoS Comput Biol 4(2):e43
Thirumal Kumar D, George Priya Doss C, Sneha P, et al (2016) Influence of V54M mutation in giant muscle protein titin: a computational screening and molecular dynamics approach. J Biomol Struct Dyn 1–12. doi: https://10.1080/07391102.2016.1166456
Vacic V, Markwick PR, Oldfield CJ, Zhao X, Haynes C, Uversky VN, Iakoucheva LM (2012) Disease-associated mutations disrupt functionally important regions of intrinsic protein disorder. PLoS Comput Biol 8:e1002709
Yun S, Guy HR (2011) Stability tests on known and misfolded structures with discrete and all atom molecular dynamics simulations. J Mol Graph Model. https://doi.org/10.1016/j.jmgm.2010.12.002
Zaki OK, Krishnamoorthy N, El Abd HS, Harche SA, Mattar RA, Al Disi RS, Nofal MY, El Bekay R, Ahmed KA, George Priya Doss C, Zayed H (2017a) Two patients with Canavan disease and structural modeling of a novel mutation. Metab Brain Dis 32(1):171–177. https://doi.org/10.1007/s11011-016-9896-9
Zaki OK, Priya Doss CG, Ali SA, Murad GG, Elashi SA, Ebnou MS, Thirumal Kumar D, Khalifa O, Gamal R, El Abd HSA, Nasr BN, Zayed H (2017b) Genotype-phenotype correlation in patients with Isovaleric Acidemia: comparative structural modelling and computational analysis of novel variants. Hum Mol Genet. https://doi.org/10.1093/hmg/ddx195
Zimmermann MT, Urrutia R, Oliver GR, Blackburn PR, Cousin MA, Bozeck NJ, Klee EW (2017) Molecular modeling and molecular dynamic simulation of the effects of variants in the TGFBR2 kinase domain as a paradigm for interpretation of variants obtained by next generation sequencing. PLoS One 12(2):e0170822
Zoref E, De Vries A, Sperling O (1975) Mutant feedback-resistant phosphoribosylpyrophosphate synthetase associated with purine over production and gout Phosphoribosylpyrophosphate and purine metabolism in cultured fibroblasts. J Clin Invest 56:1093–1099
Acknowledgements
We would like to thank VIT University and CDAC@BRAF for providing the facility. Ashish Kumar Agrahari is extremely grateful to Dr. Mitali Mukerji (Senior Principal Scientist at the CSIR Institute of Genomics and Integrative Biology) for the providing the facility at CSIR-IGIB to carrying out the project in generating the secondary structure bar plot (Fig. 1).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
All authors declare no conflict of interest.
Electronic supplementary material
ESM 1
(DOCX 2084 kb)
Rights and permissions
About this article
Cite this article
Agrahari, A.K., Sneha, P., George Priya Doss, C. et al. A profound computational study to prioritize the disease-causing mutations in PRPS1 gene. Metab Brain Dis 33, 589–600 (2018). https://doi.org/10.1007/s11011-017-0121-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11011-017-0121-2