Abstract
DNA methyltransferases (DNMTs) represent promising targets for the development of unique anticancer drugs. However, all DNMT inhibitors currently in clinical use are nonselective cytosine analogs with significant cytotoxic side-effects. Several natural products, covering diverse chemical classes, have indicated DNMT inhibitory activity, but these effects have yet to be systematically evaluated. In this study, we provide experimental data suggesting that two of the most prominent natural products associated with DNA methylation inhibition, (−)-epigallocathechin-3-gallate (EGCG) and curcumin, have little or no pharmacologically relevant inhibitory activity. We therefore conducted a virtual screen of a large database of natural products with a validated homology model of the catalytic domain of DNMT1. The virtual screening focused on a lead-like subset of the natural products docked with DNMT1, using three docking programs, following a multistep docking approach. Prior to docking, the lead-like subset was characterized in terms of chemical space coverage and scaffold content. Consensus hits with high predicted docking affinity for DNMT1 by all three docking programs were identified. One hit showed DNMT1 inhibitory activity in a previous study. The virtual screening hits were located within the biological-relevant chemical space of drugs, and represent potential unique DNMT inhibitors of natural origin. Validation of these virtual screening hits is warranted.
Similar content being viewed by others
Abbreviations
- DNMT:
-
DNA methyltransferase
- EGCG:
-
(−)-epigallocathechin-3-gallate
- HBA:
-
Hydrogen bond acceptor
- HBD:
-
Hydrogen bond donor
- MOE:
-
Molecular operating environment
- MW:
-
Molecular weight
- NCI:
-
National Cancer Institute
- RB:
-
Rotatable bond
- SAH:
-
S-adenosyl-l-homocysteine
- SAM:
-
S-adenosyl-l-methionine
- SP:
-
Standard precision
- TPSA:
-
Topological polar surface area
- XP:
-
Extra precision
References
Goll MG, Bestor TH (2005) Eukaryotic cytosine methyltransferases. Annu Rev Biochem 74: 481–514. doi:10.1146/annurev.biochem.74.010904.153721
Jones PA, Baylin SB (2007) The epigenomics of cancer. Cell 128: 683–692. doi:10.1016/j.cell.2007.01.029
Lyko F, Brown R (2005) DNA methyltransferase inhibitors and the development of epigenetic cancer therapies. J Natl Cancer Inst 97: 1498–1506. doi:10.1093/jnci/dji311
Brueckner B, Kuck D, Lyko F (2007) DNA methyltransferase inhibitors for cancer therapy. Cancer J 13: 17–22. doi:10.1097/PPO.0b013e31803c7245
Santi DV, Garrett CE, Barr PJ (1983) On the mechanism of inhibition of DNA cytosine methyltransferases by cytosine analogs. Cell 33: 9–10. doi:10.1016/0092-8674(83)90327-6
Wu JC, Santi DV (1987) Kinetic and catalytic mechanism of HhaI methyltransferase. J Biol Chem 262: 4778–4786
Chen L, Macmillan AM, Chang W, Ezaznikpay K, Lane WS, Verdine GL (1991) Direct identification of the active-site nucleophile in a DNA (cytosine-5)-methyltransferase. Biochemistry 30: 11018–11025. doi:10.1021/bi00110a002
Ogara M, Klimasauskas S, Roberts RJ, Cheng XD (1996) Enzymatic C5-cytosine methylation of DNA: mechanistic implications of new crystal structures for HhaI methyltransferase-DNA-AdoHcy complexes. J Mol Biol 261: 634–645. doi:10.1006/jmbi.1996.0489
Kumar S, Horton JR, Jones GD, Walker RT, Roberts RJ, Cheng X (1997) DNA containing 4’-thio-2’-deoxycytidine inhibits methylation by HhaI methyltransferase. Nucleic Acids Res 25: 2773–2783. doi:10.1093/nar/25.14.2773
Jurkowski TP, Meusburger M, Phalke S, Helm M, Nellen W, Reuter G, Jeltsch A (2008) Human DNMT2 methylates tRNA(Asp) molecules using a DNA methyltransferase-like catalytic mechanism. RNA 14: 1663–1670. doi:10.1261/rna.970408
Suzuki T, Miyata N (2006) Epigenetic control using natural products and synthetic molecules. Curr Med Chem 13: 935–958. doi:10.2174/092986706776361067
Liu K, Wang YF, Cantemir C, Muller MT (2003) Endogenous assays of DNA methyltransferases: evidence for differential activities of DNMT1, DNMT2, and DNMT3 in mammalian cells in vivo. Mol Cell Biol 23: 2709–2719. doi:10.1128/mcb.23.8.2709-2719.2003
Stresemann C, Lyko F (2008) Modes of action of the DNA methyltransferase inhibitors azacytidine and decitabine. Int J Cancer 123: 8–13. doi:10.1002/ijc.23607
Schermelleh L, Spada F, Easwaran HP, Zolghadr K, Margot JB, Cardoso MC, Leonhardt H (2005) Trapped in action: direct visualization of DNA methyltransferase activity in living cells. Nat Methods 2: 751–756. doi:10.1038/nmeth794
Segura-Pacheco B, Trejo-Becerril C, Perez-Cardenas E, Taja-Chayeb L, Mariscal I, Chavez A, Acuna C, Salazar AM, Lizano M, Duenas-Gonzalez A (2003) Reactivation of tumor suppressor genes by the cardiovascular drugs hydralazine and procainamide and their potential use in cancer therapy. Clin Cancer Res 9: 1596–1603
Segura-Pacheco B, Perez-Cardenas E, Taja-Chayeb L, Chavez-Blanco A, Revilla-Vazquez A, Benitez-Bribiesca L, Duenas-Gonzalez A (2006) Global DNA hypermethylation-associated cancer chemotherapy resistance and its reversion with the demethylating agent hydralazine. J Transl Med 4: 32. doi:10.1186/1479-5876-4-32
Villar-Garea A, Fraga MF, Espada J, Esteller M (2003) Procaine is a DNA-demethylating agent with growth-inhibitory effects in human cancer cells. Cancer Res 63: 4984–4989
Lee BH, Yegnasubramanian S, Lin XH, Nelson WG (2005) Procainamide is a specific inhibitor of DNA methyltransferase 1. J Biol Chem 280: 40749–40756. doi:10.1074/jbc.M505593200
Castellano S, Kuck D, Sala M, Novellino E, Lyko F, Sbardella G (2008) Constrained analogues of procaine as novel small molecule inhibitors of DNA methyltransferase-1. J Med Chem 51: 2321–2325. doi:10.1021/jm7015705
Siedlecki P, Boy RG, Musch T, Brueckner B, Suhai S, Lyko F, Zielenkiewicz P (2006) Discovery of two novel, small-molecule inhibitors of DNA methylation. J Med Chem 49: 678–683. doi:10.1021/jm050844z
Kuck D, Singh N, Lyko F, Medina-Franco JL (2010) Novel and selective DNA methyltransferase inhibitors: docking-based virtual screening and experimental evaluation. Bioorg Med Chem 18: 822–829. doi:10.1016/j.bmc.2009.11.050
Harvey AL (2008) Natural products in drug discovery. Drug Discov Today 13: 894–901. doi:10.1016/j.drudis.2008.07.004
Ganesan A (2008) The impact of natural products upon modern drug discovery. Curr Opin Chem Biol 12: 306–317. doi:10.1016/j.cbpa.2008.03.016
Butler MS (2008) Natural products to drugs: natural product-derived compounds in clinical trials. Nat Prod Rep 25: 475–516. doi:10.1039/b514294f
Newman DJ (2008) Natural products as leads to potential drugs: an old process or the new hope for drug discovery?. J Med Chem 51: 2589–2599. doi:10.1021/jm0704090
Molinski TF, Dalisay DS, Lievens SL, Saludes JP (2009) Drug development from marine natural products. Nat Rev Drug Discov 8: 69–85. doi:10.1038/nrd2487
Demain AL, Sanchez S (2009) Microbial drug discovery: 80 years of progress. J Antibiot 62: 5–16. doi:10.1038/ja.2008.16
Cragg GM, Newman DJ (2009) Nature: a vital source of leads for anticancer drug development. Phytochem Rev 8: 313–331. doi:10.1007/s11101-009-9123-y
Coseri S (2009) Natural products and their analogues as efficient anticancer drugs. Mini-Rev Med Chem 9: 560–571. doi:10.2174/138955709788167592
Kinghorn AD, Chin YW, Swanson SM (2009) Discovery of natural product anticancer agents from biodiverse organisms. Curr Opin Drug Discov Dev 12: 189–196
Hauser AT, Jung M (2008) Targeting epigenetic mechanisms: potential of natural products in cancer chemoprevention. Planta Med 74: 1593–1601. doi:10.1055/s-2008-1081347
Yu N, Wang M (2008) Anticancer drug discovery targeting DNA hypermethylation. Curr Med Chem 15: 1350–1375. doi:10.2174/092986708784567653
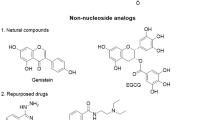
Fang MZ, Wang YM, Ai N, Hou Z, Sun Y, Lu H, Welsh W, Yang CS (2003) Tea polyphenol (−)-epigallocatechin-3-gallate inhibits DNA methyltransferase and reactivates methylation-silenced genes in cancer cell lines. Cancer Res 63: 7563–7570
Lee WJ, Shim JY, Zhu BT (2005) Mechanisms for the inhibition of DNA methyltransferases by tea catechins and bioflavonoids. Mol Pharmacol 68: 1018–1030. doi:10.1124/mol.104.008367
Lee WJ, Zhu BT (2006) Inhibition of DNA methylation by caffeic acid and chlorogenic acid, two common catechol-containing coffee polyphenols. Carcinogenesis 27: 269–277. doi:10.1093/carcin/bgi206
Fini L, Selgrad M, Fogliano V, Graziani G, Romano M, Hotchkiss E, Daoud YA, De Vol EB, Boland CR, Ricciardiello L (2007) Annurca apple polyphenols have potent demethylating activity and can reactivate silenced tumor suppressor genes in colorectal cancer cells. J Nutr 137: 2622–2628. doi:10.3945/jn.109.118521
Fang MZ, Chen DP, Sun Y, Jin Z, Christman JK, Yang CS (2005) Reversal of hypermethylation and reactivation of p16INK4a, RARβ, and MGMT genes by genistein and other isoflavones from soy. Clin Cancer Res 11: 7033–7041. doi:10.1158/1078-0432.ccr-05-0406
Jagadeesh S, Sinha S, Pal BC, Bhattacharya S, Banerjee PP (2007) Mahanine reverses an epigenetically silenced tumor suppressor gene RASSF1A in human prostate cancer cells. Biochem Biophys Res Commun 362: 212–217. doi:10.1016/j.bbrc.2007.08.005
Pina IC, Gautschi JT, Wang GYS, Sanders ML, Schmitz FJ, France D, Cornell-Kennon S, Sambucetti LC, Remiszewski SW, Perez LB, Bair KW, Crews P (2003) Psammaplins from the sponge Pseudoceratina purpurea: inhibition of both histone deacetylase and DNA methyltransferase. J Org Chem 68: 3866–3873. doi:10.1021/jo034248t
McPhail KL, France D, Cornell-Kennon S, Gerwick WH (2004) Peyssonenynes A and B, novel enediyne oxylipins with DNA methyl transferase inhibitory activity from the red marine alga Peyssonnelia caulifera. J Nat Prod 67: 1010–1013. doi:10.1021/np0400252
Liu ZF, Liu SJ, Xie ZL, Pavlovicz RE, Wu J, Chen P, Aimiuwu J, Pang JX, Bhasin D, Neviani P, Fuchs JR, Plass C, Li PK, Li C, Huang THM, Wu LC, Rush L, Wang HY, Perrotti D, Marcucci G, Chan KK (2009) Modulation of DNA methylation by a sesquiterpene lactone parthenolide. J Pharmacol Exp Ther 329: 505–514. doi:10.1124/jpet.108.147934
Liu ZF, Xie ZL, Jones W, Pavlovicz RE, Liu SJ, Yu JH, Li PK, Lin JY, Fuchs JR, Marcucci G, Li CL, Chan KK (2009) Curcumin is a potent DNA hypomethylation agent. Bioorg Med Chem Lett 19: 706–709. doi:10.1016/j.bmcl.2008.12.041
Singh N, Dueñas-González A, Lyko F, Medina-Franco JL (2009) Molecular modeling and dynamics studies of hydralazine with human DNA methyltransferase 1. ChemMedChem 4: 792–799. doi:10.1002/cmdc.200900017
Siedlecki P, Boy RG, Comagic S, Schirrmacher R, Wiessler M, Zielenkiewicz P, Suhai S, Lyko F (2003) Establishment and functional validation of a structural homology model for human DNA methyltransferase 1. Biochem Biophys Res Commun 306: 558–563. doi:10.1016/s0006-291x(03)01000-3
Brueckner B, Boy RG, Siedlecki P, Musch T, Kliem HC, Zielenkiewicz P, Suhai S, Wiessler M, Lyko F (2005) Epigenetic reactivation of tumor suppressor genes by a novel small-molecule inhibitor of human DNA methyltransferases. Cancer Res 65: 6305–6311. doi:10.1158/0008-5472.CAN-04-2957
Stach D, Schmitz OJ, Stilgenbauer S, Benner A, Dohner H, Wiessler M, Lyko F (2003) Capillary electrophoretic analysis of genomic DNA methylation levels. Nucleic Acids Res 31: e2. doi:10.1093/nar/gng002
Irwin JJ, Shoichet BK (2005) ZINC—a free database of commercially available compounds for virtual screening. J Chem Inf Model 45: 177–182. doi:10.1021/ci049714+
FILTER, version 2.0.2. OpenEye Scientific Software Inc., Santa Fe, NM. http://www.eyesopen.com. Accessed April 2010)
LigPrep, version 2.2 (2005) Schrödinger, LLC, New York, NY
Xu Y, Johnson M (2001) Algorithm for naming molecular equivalence classes represented by labeled pseudographs. J Chem Inf Comput Sci 41: 181–185. doi:10.1021/ci0003911
Xu YJ, Johnson M (2002) Using molecular equivalence numbers to visually explore structural features that distinguish chemical libraries. J Chem Inf Comput Sci 42: 912–926. doi:10.1021/ci025535l
Xu J (2002) A new approach to finding natural chemical structure classes. J Med Chem 45: 5311–5320. doi:10.1021/jm010520k
Liu B, Lu A, Zhang L, Liu H, Liu Z, Zhou J (2004) New diversity criterion and database compression method. Internet Electron J Mol Des 3:143–149. http://www.biochempress.com
Bemis GW, Murcko MA (1996) The properties of known drugs. 1. Molecular frameworks. J Med Chem 39: 2887–2893. doi:10.1021/jm9602928
Medina-Franco JL, Petit J, Maggiora GM (2006) Hierarchical strategy for identifying active chemotype classes in compound databases. Chem Biol Drug Des 67: 395–408. doi:10.1111/j.1747-0285.2006.00397.x
Medina-Franco JL, Martinez-Mayorga K, Giulianotti MA, Houghten RA, Pinilla C (2008) Visualization of the chemical space in drug discovery. Curr Comput Aided Drug Des 4: 322–333. doi:10.2174/157340908786786010
Singh N, Guha R, Giulianotti MA, Pinilla C, Houghten RA, Medina-Franco JL (2009) Chemoinformatic analysis of combinatorial libraries, drugs, natural products, and molecular libraries small molecule repository. J Chem Inf Model 49: 1010–1024. doi:10.1021/ci800426u
Medina-Franco JL, Martínez-Mayorga K, Bender A, Scior T (2009) Scaffold diversity analysis of compound data sets using an entropy-based measure. QSAR Comb Sci 28: 1551–1560. doi:10.1002/qsar.200960069
Molecular Operating Environment (MOE), version 2008.10. Chemical Computing Group Inc., Montreal, QC, Canada. http://www.chemcomp.com. Accessed April 2010
Glide, version 5.0 (2008) Schrödinger, LLC, New York, NY
Jones G, Willett P, Glen RC, Leach AR, Taylor R (1997) Development and validation of a genetic algorithm for flexible docking. J Mol Biol 267: 727–748. doi:10.1006/jmbi.1996.0897
Huey R, Morris GM, Olson AJ, Goodsell DS (2007) A semiempirical free energy force field with charge-based desolvation. J Comput Chem 28: 1145–1152. doi:10.1002/jcc.20634
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30: 2785–2791. doi:10.1002/jcc.21256
Gasteiger J, Marsili M (1980) Iterative partial equalization of orbital electronegativity—a rapid access to atomic charges. Tetrahedron 36: 3219–3228. doi:10.1016/0040-4020(80)80168-2
Chuang JC, Yoo CB, Kwan JM, Li TWH, Liang GN, Yang AS, Jones PA (2005) Comparison of biological effects of non-nucleoside DNA methylation inhibitors versus 5-aza-2’-deoxycytidine. Mol Cancer Ther 4: 1515–1520. doi:10.1158/1535-7163.MCT-05-0172
Stresemann C, Brueckner B, Musch T, Stopper H, Lyko F (2006) Functional diversity of DNA methyltransferase inhibitors in human cancer cell lines. Cancer Res 66: 2794–2800. doi:10.1158/0008-5472.CAN-05-2821
Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, Gautam B, Hassanali M (2008) DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res 36: D901–D906. doi:10.1093/nar/gkm958
Charifson PS, Walters WP (2002) Filtering databases and chemical libraries. J Comput Aided Mol Des 16: 311–323. doi:10.1023/A:1020829519597
Talevi A, Gavernet L, Bruno-Blanch LE (2009) Combined virtual screening strategies. Curr Comput Aided Drug Des 5: 23–37. doi:10.2174/157340909787580854
Musa MA, Cooperwood JS, Khan MOF (2008) A review of coumarin derivatives in pharmacotherapy of breast cancer. Curr Med Chem 15: 2664–2679
Wu L, Wang X, Xu W, Farzaneh F, Xu R (2009) The structure and pharmacological functions of coumarins and their derivatives. Curr Med Chem 16: 4236–4260. doi:10.2174/092986709789578187
CRC Dictionary of Natural Products. http://www.crcpress.com/. Accessed April 2010
Drug Discovery Portal. http://www.ddp.strath.ac.uk/. Accessed April 2010
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
The Below is the Electronic Supplementary Material.
Rights and permissions
About this article
Cite this article
Medina-Franco, J.L., López-Vallejo, F., Kuck, D. et al. Natural products as DNA methyltransferase inhibitors: a computer-aided discovery approach. Mol Divers 15, 293–304 (2011). https://doi.org/10.1007/s11030-010-9262-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11030-010-9262-5