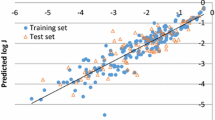
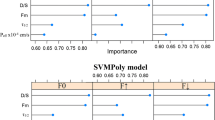
Abstract
In many absorption, distribution, metabolism, and excretion (ADME) modeling problems, imbalanced data could negatively affect classification performance of machine learning algorithms. Solutions for handling imbalanced dataset have been proposed, but their application for ADME modeling tasks is underexplored. In this paper, various strategies including cost-sensitive learning and resampling methods were studied to tackle the moderate imbalance problem of a large Caco-2 cell permeability database. Simple physicochemical molecular descriptors were utilized for data modeling. Support vector machine classifiers were constructed and compared using multiple comparison tests. Results showed that the models developed on the basis of resampling strategies displayed better performance than the cost-sensitive classification models, especially in the case of oversampling data where misclassification rates for minority class have values of 0.11 and 0.14 for training and test set, respectively. A consensus model with enhanced applicability domain was subsequently constructed and showed improved performance. This model was used to predict a set of randomly selected high-permeability reference drugs according to the biopharmaceutics classification system. Overall, this study provides a comparison of numerous rebalancing strategies and displays the effectiveness of oversampling methods to deal with imbalanced permeability data problems.


Similar content being viewed by others
Abbreviations
- AD:
-
Applicability domain
- ADME:
-
Absorption, distribution, metabolism, and excretion
- AURC:
-
Area under the ROC
- BCS:
-
Biopharmaceutics classification system
- BE:
-
Bioequivalence
- C :
-
Penalty parameter
- CD:
-
Critical distance
- EMA:
-
European medicines agency
- F:
-
Bioavailability
- FN:
-
False negative
- FDA:
-
US Food and Drug Administration
- FP:
-
False positive
- H class:
-
High-permeability class
- HIA:
-
Human intestinal absorption
- IVIVC:
-
In vitro–In vivo correlation
- MD:
-
Molecular descriptor
- M-P class:
-
Moderate-to-poor permeability class
- Papp:
-
Apparent permeability coefficient
- RBF:
-
Radial basis function
- ROC:
-
Receiver operator curve
- SMOTE:
-
Synthetic minority oversampling technique
- SVM:
-
Support vector machine
- SVs:
-
Support vectors
- WHO:
-
World health organization
References
Chawla NV (2010) Data mining for imbalanced datasets: an overview. In: Data mining and knowledge discovery handbook. Maimon O, Rokach L (eds). vol 45, 2nd edn. Springer, 233 Spring Street, New York, NY 10013, USA, pp 875–886. doi:10.1007/978-0-387-09823-4
Japkowicz N (2003) Class imbalances: are we focusing on the right issue? Paper presented at the ICML’2003 Workshop on learning from imbalanced data sets (II). Washington, DC, 21 August 2003
Drummond C, Holte RC (2003) C4.5, Class imbalance, and cost sensitivity: why under-sampling beats over-sampling. In: Proceedings of the international conference on machine learning (ICML 2003) Workshop on learning from imbalanced data sets II, Washington, DC
Trotter MWB, Holden SB (2003) Support vector machines for ADME property classification. QSAR Comb Sci 22:533–548. doi:10.1002/qsar.200310006
Pinto M, Trauner M, Ecker GF (2012) An in silico classification model for putative ABCC2 substrates. Mol Inf 31:547–553. doi:10.1002/minf.201200049
Eitrich T, Kless A, Druska C, Meyer B, Grotendorst J (2007) Classification of highly unbalanced CYP450 data of drugs using cost sensitive machine learning techniques. J Chem Inf Model 47:92–103. doi:10.1021/ci6002619
Hou T, Wang J, Li Y (2007) ADME evaluation in drug discovery. 8. The prediction of human intestinal absorption by a support vector machine. J Chem Inf Model 47:2408–2415. doi:10.1021/ci7002076
Newby D, Freitas AA, Ghafourian T (2013) Coping with unbalanced class data sets in oral absorption models. J Chem Inf Model 53:461–474. doi:10.1021/ci300348u
Avdeef A (2003) Absorption and drug development: solubility, permeability, and charge state, 1st edn. Wiley, Hoboken. doi:10.1002/047145026X
Oltra-Noguera D, Mangas-Sanjuan V, Centelles-Sangüesa A, Gonzalez-Garcia I, Sanchez-Castaño G, Gonzalez-Alvarez M, Casabo V-G, Merino V, Gonzalez-Alvarez I, Bermejo M (2015) Variability of permeability estimation from different protocols of subculture and transport experiments in cell monolayers. J Pharmacol Toxicol Methods 71:21–32. doi:10.1016/j.vascn.2014.11.004
Pham-The H, Garrigues T, Bermejo M, González-Álvarez I, Monteagudo MC, Cabrera-Pérez MÁ (2013) Provisional classification and in silico study of biopharmaceutical system based on Caco-2 cell permeability and dose number. Mol Pharm 10:2445–2461. doi:10.1021/mp4000585
Pham-The H, González-Álvarez I, Bermejo M, Garrigues T, Le-Thi-Thu H, Cabrera-Pérez MÁ (2013) The use of rule-based and QSPR approaches in ADME profiling: a case study on Caco-2 permeability. Mol Inf 32:459–479. doi:10.1002/minf.201200166
Annex 8: Proposal to waive in vivo bioequivalence requirements for WHO Model List of Essential Medicines immediate-release, solid oral dosage forms (2006) WHO Expert Committee on specification for pharmaceutical preparations. WHO Technical Report Series No. 937:391-461. http://www.who.int/medicines/publications/essentialmedicines/en/index.html
CDER/FDA FDA Guidance for industry: waiver of in vivo bioavailability and bioequivalence studies for immediate-release solid oral dosage forms based on a biopharmaceutics classification system (2000) Federal Drug and Food Administration, Rockville. www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/ucm070246.pdf
Pham-The H, Gonzalez-Diaz I, Bermejo-Sanz M, Mangas-Sanjuan V, Centelles I, Garriges TM, Cabrera-Perez MA (2011) In silico prediction of Caco-2 permeability by a classification QSAR approach. Mol Inf 30:376–385. doi:10.1002/minf.201000118
Le-Thi-Thu H, Canizares-Carmenate Y, Marrero-Ponce Y, Torrens F, Castillo-Garit JA (2015) Prediction of Caco-2 cell permeability using bilinear indices and multiple linear regression. Lett Drug Des Discov, vol 12 (E-pub ahead of print). doi:10.2174/1570180812666150630183511
Prieto P, Hoffmann S, Tirelli V, Tancredi F, González I, Bermejo M, De Angelis I (2010) An exploratory study of two Caco-2 cell models for oral absorption: a report on their within-laboratory and between-laboratory variability, and their predictive capacity. Altern Lab Anim 38:367–386
Volpe DA (2008) Variability in Caco-2 and MDCK cell-based intestinal permeability assays. J Pharm Sci 97:712–725. doi:10.1002/jps.21010
Polli JE, Yu LX, Cook JA, Amidon GL, Borchardt RT, Burnside BA, Burton PS, Chen ML, Conner DP, Faustino PJ, Hawi AA, Hussain AS, Joshi HN, Kwei G, Lee VH, Lesko LJ, Lipper RA, Loper AE, Nerurkar SG, Polli JW, Sanvordeker DR, Taneja R, Uppoor RS, Vattikonda CS, Wilding I, Zhang G (2004) Summary workshop report: biopharmaceutics classification system-implementation challenges and extension opportunities. J Pharm Sci 93:1375–1381. doi:10.1002/jps.20064
Kim JS, Mitchell S, Kijek P, Tsume Y, Hilfinger J, Amidon GL (2006) The suitability of an in situ perfusion model for permeability determinations: utility for BCS Class I biowaiver requests. Mol Pharm 3:686–694. doi:10.1021/mp060042f
Maenner MJ, Denlinger LC, Langton A, Meyers KJ, Engelman CD, Skinner HG (2009) Detecting gene-by-smoking interactions in a genome-wide association study of early-onset coronary heart disease using random forests. BMC Proc 3(Suppl 7):S88. doi:10.1186/1753-6561-3-S7-S88
HyperChem (TM) Professional 8.0.5. Hypercube, Inc., 1115 NW 4th Street, Gainesville, Florida 32601, USA. (www.hyper.com/)
STATISTICA (data analysis software system) (2007). 8.0 edn. StatSoft, Inc., Tulsa. (www.statsoft.com)
Vapnik V (1995) The nature of statistical learning theory. Springer, New York
Burges CJC (1998) A tutorial on support vector machines for pattern recognition. Data Min Knowl Disc 2:127–167. doi:10.1234/12345678
Chang C-C, Lin C-J (2011) LIBSVM: a library for support vector machines. ACM Trans Intell Syst Technol 2:1–27. doi:10.1145/1961189.1961199
Hsu C-W, Chang C-C, Lin C-J (2003) A practical guide to support vector classification. Department of Computer Science, National Taiwan University, http://www.csie.ntu.edu.tw/~cjlin. Accessed 17 October 2014
Platt JC (1999) Fast training of support vector machines using sequential minimal optimization. Advances in kernel methods. MIT Press, Cambridge, pp 185–208
Witten HI, Frank E (2005) Data mining: practical machine learning tools and techniques, 2nd edn. Morgan Kaufmann, San Francisco
Akbani R, Kwek S, Japkowicz N (2004) Applying support vector machines to imbalanced datasets. In: Boulicaut J-F, Esposito F, Giannotti F, Pedreschi D (eds) Machine learning: ECML 2004, vol 3201., Lecture notes in computer science. Springer, Berlin, pp 39–50. doi:10.1007/978-3-540-30115-8_7
Wang BX, Japkowicz N (2010) Boosting support vector machines for imbalanced data sets. Knowl Inf Syst 25:1–20. doi:10.1007/s10115-009-0198-y
Veropoulos K, Campbell C, Cristianini N (1999) Controlling the sensitivity of support vector machines. In: International joint conference on AI (IJCAI 99), Stockholm, pp 55–60
Wu G, Chang EY (2003) Adaptive feature-space conformal transformation for imbalanced-data learning. In: Proceeding of the 20th international conference on machine learning (ICML-2003), vol 2. Washington DC, pp 816–823
Schierz AC (2009) Virtual screening of bioassay data. J Cheminform 1:1–12. doi:10.1186/1758-2946-1-21
Domingos P (1999) MetaCost: A general method for making classifiers cost-sensitive. In: KDD ’99 Proceedings of the 5th ACM SIGKDD international conference on Knowledge discovery and data mining. ACM, San Diego, pp 155–164, doi:10.1145/312129.312220
Jo T, Japkowicz N (2004) Class imbalances versus small disjuncts. ACM SIGKDD Explor Newsl 6:40–49. doi:10.1145/1007730.1007737
Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP (2002) Synthetic minority over-sampling technique. J Artif Intell Res 16:321–357. doi:10.1613/jair.953
Kohavi R, John GH (1997) Wrappers for feature subset selection. Artif Intell 97:273–324. doi:10.1016/S0004-3702(97)00043-X
John GH, Kohavi R, Pfleger K (1994) Irrelevant features and the subset selection problem. In: Cohen WW, Hirsh H (eds) Machine learning proceedings of the eleventh international conference. Morgan Kaufmann, San Francisco, pp 121–129
Jaworska J, Nikolova-Jeliazkova N, Aldenberg T (2005) QSAR applicabilty domain estimation by projection of the training set descriptor space: a review. Altern Lab Anim 33:445–459
Provost F, Fawcett T (1997) Analysis and visualization of classifier performance: comparison under imprecise class and cost distributions. In: Proceedings of the 3rd international conference on knowledge discovery and data mining (KDD-97), Newportbeach, August 1997, pp 43–48
Le-Thi-Thu H, Casanola-Martín GM, Marrero-Ponce Y, Rescigno A, Abad C, Khan MT (2014) A rational workflow for sequential virtual screening of chemical libraries on searching for new tyrosinase inhibitors. Curr Top Med Chem 14:1473–1485. doi:10.2174/1568026614666140523120336
Demsar J (2006) Statistical comparisons of classifiers over multiple data sets. J Mach Learn Res 7:1–30
Le-Thi-Thu H, Marrero-Ponce Y, Casanola-Martin GM, Cardoso GC, Chávez MC, Garcia MM, Morell C, Torrens F, Abad C (2011) A comparative study of nonlinear machine learning for the “In silico” depiction of Tyrosinase Inhibitory Activity from Molecular Structure. Mol Inf 30:527–537. doi:10.1002/minf.201100021
Friedman M (1940) A comparison of alternative tests of significance for the test of m rankings. Ann math Statist 11:86–92. doi:10.2307/2235971
Iman RL, Davenport JM (1980) Approximations of the critical region of the Friedman statistic. Commun Stat 9:571–595. doi:10.1080/03610928008827904
Dunn OJ (1961) Multiple comparisons among means. J Am Stat Assoc 56:52–64. doi:10.2307/2282330
Le-Thi-Thu H, Cardoso GC, Casañola-Martin GM, Marrero-Ponce Y, Puris A, Torrens F, Rescigno A, Abad A (2010) QSAR models for tyrosinase inhibitory activity description applying modern statistical classification techniques: A comparative study. Chemom Intell Lab Syst 104:249–259. doi:10.1016/j.chemolab.2010.08.016
Fawcett T (2003) ROC Graphs: notes and practical considerations for data mining researchers. Technical Report HPL-2003-4. HP Laboratories, Palo Alto
Oprea T (2000) Property distribution of drug-related chemical databases. J Comput Aided Mol Des 14:251–264. doi:10.1023/A:1008130001697
Congreve M, Carr R, Murray C, Jhoti H (2003) A rule of three for fragment: based lead discovery? Drug Discov Today 8:876–877. doi:10.1016/S1359-6446(03)02831-9
Cabrera-Perez MA, Pham-The H, Bermejo M, Alvarez IG, Alvarez MG, Garrigues TM (2012) QSPR in oral bioavailability: specificity or integrality? Mini-Rev Med Chem 12:534–550. doi:10.2174/138955712800493753
Tremblay P, Auger S, Picard P, Blachon G, Julian B, Laplanche L, Sarcy C, Estoul S, Moliner P, Fedeli O, Fabre G (2010) LDTD384-MS/MS for in vitro assays. Paper presented at the 58th ASMS Conference on Mass Spectrometry, Salt Lake City
Hu M, Ling J, Lin H, Chen J (2004) Use of Caco-2 cell monolayers to study drug absorption and metabolism. In: Yan Z, Caldwell GW (eds) Optimization in drug discovery: in vitro methods, vol 2., Methods in pharmacology and toxicologyHumana Press Inc., Totowa, pp 19–35. doi:10.1385/1-59259-800-5:019
Dressman JB, Nair A, Abrahamsson B, Barends DM, Groot DW, Kopp S, Langguth P, Polli JE, Shah VP, Zimmer M (2012) Biowaiver monograph for immediate-release solid oral dosage forms: acetylsalicylic acid. J Pharm Sci 101:2653–2667. doi:10.1002/jps.23212
Letcher SG (2010) Phylogenetic structure of angiosperm communities during tropical forest succession. Proc Biol Sci 277:97–104. doi:10.1098/rspb.2009.0865
Zhao YH, Le J, Abraham MH, Hersey A, Eddershaw PJ, Luscombe CN, Butina D, Beck G, Sherborne B, Cooper I, Platts JA (2001) Evaluation of human intestinal absorption data and subsequent derivation of a quantitative structure-activity relationship (QSAR) with the Abraham descriptors. J Pharm Sci 90:749–784. doi:10.1002/jps.1031
Butler JM, Dressman JB (2010) The developability classification system: application of biopharmaceutics concepts to formulation development. J Pharm Sci 99:4940–4954. doi:10.1002/jps.22217
Acknowledgments
H.L-T-T is supported by Vietnam National University. H.P-T, M.B, I.G-A, T.G, and M.A.C-P acknowledge financial support of AECID (Grant No. 1- D/031152/10 and DCI-ALA/19.09.01/10/21526/245-297/ALFA 111(2010)29). We greatly appreciate Mr. Aaron Burns from Oxford English UK Vietnam for his careful review and helpful editing of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Pham-The, H., Casañola-Martin, G., Garrigues, T. et al. Exploring different strategies for imbalanced ADME data problem: case study on Caco-2 permeability modeling. Mol Divers 20, 93–109 (2016). https://doi.org/10.1007/s11030-015-9649-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11030-015-9649-4