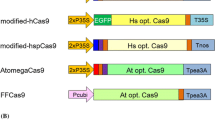
Abstract
CRISPR/Cas9 is a novel tool for targeted mutagenesis and is applicable to plants, including rice. Previous reports on CRISPR/Cas9 in rice have demonstrated that target mutations are transmitted to the next generation in accordance with Mendelian law, but heritability of the target mutation and the role of inherited Cas9 gene have not been fully elucidated. Here, we targeted the rice phytoene desaturase gene, mutants of which exhibit an albino phenotype, by using CRISPR/Cas9 and analyzed segregation of target mutations. Agrobacterium-mediated methods using immature embryos successfully transformed a CRISPR/Cas9 system into five rice cultivars and subsequently induced mutation. Unpredicted segregations, with more mutants than theoretically predicted, were frequently found in T1 plants from monoallelic T0 mutants. Chimeric plants with both biallelic and monoallelic mutated cells were also observed in the T1. Next, we followed segregation of a target mutation in the T2 from monoallelic T1 mutants. When T1 mutants possessed Cas9, unpredicted segregations of the target mutation and chimeric plants were observed again in the T2. When T1 mutants did not possess Cas9, segregation of the target mutations followed Mendelian law and no chimeric plants appeared in the T2. T2 mutants with Cas9 had mutations different from the original ones found in T0. Our results indicated that inherited Cas9 was still active in later generations and could induce new mutations in the progeny, leading to chimerism and unpredicted segregation. We conclude that Cas9 has to be eliminated by segregation in T1 to generate homozygous mutants without chimerism or unpredicted segregation.





Similar content being viewed by others
References
Baysal C, Bortesi L, Zhu C, Farré G, Schillberg S, Christou P (2016) CRISPR/Cas9 activity in the rice OsBEIIb gene does not induce off-target effects in the closely related paralog OsBEIIa. Mol Breeding 36:1–11. doi:10.1007/s11032-016-0533-4
Belhaj K, Chaparro-Garcia A, Kamoun S, Nekrasov V (2013) Plant genome editing made easy: targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Methods 9:1–10. doi:10.1186/1746-4811-9-39
Bortesi L, Fischer R (2015) The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol Adv 33:41–52. doi:10.1016/j.biotechadv.2014.12.006
Bortesi L, Zhu C, Zischewski J, Perez L, Bassié L, Nadi R, Forni G, Lade SB, Soto E, Jin X, Medina V, Villorbina G, Muñoz P, Farré G, Fischer R, Twyman RM, Capell T, Christou P, Schillberg S (2016) Patterns of CRISPR/Cas9 activity in plants, animals and microbes. Plant Biotechnol J doi. doi:10.1111/pbi.12634
Chari R, Mali P, Moosburner M, Church GM (2015) Unraveling CRISPR-Cas9 genome engineering parameters via a library-on-library approach. Nat Methods doi. doi:10.1038/nmeth.3473
Christou P, Ford TL, Kofron M (1991) Production of transgenic rice (Oryza sativa L.) plants from agronomically important indica and japonica varieties via electric discharge particle acceleration of exogenous DNA into immature zygotic embryos. Bio-Technol 9:957–962. doi:10.1038/nbt1091-957
de Morais O, da Castro EM, Soares AA, Guimarães EP, Chatel M, Ospina Y, de Lopes AM, de Pereira JA, Utumi MM, Centeno AC, Fonseca R, Breseghello F, Guimaraes CM, Bassinello PZ, Sitarama Prabhu A, Ferreira E, Gervini de Souza NR, Alves de Souza M, Sousa Reis M, Guimaraes Santos P (2005) BRSMG Curinga: cultivar de arroz de terras altas de ampla adaptacicão para o Brasil. Embrapa Arroz e Feijão. Comunicado Técnico 114:1–8
Dellaporta SL, Wood J, Hicks JB (1983) A plant DNA minipreparation: version II. Plant Mol Biol Rep 1:19–21
Dong J, Teng W, Buchholz W, Hall T (1996) Agrobacterium-mediated transformation of Javanica rice. Mol Breeding 2:267–276. doi:10.1007/BF00564204
Endo M, Mikami M, Toki S (2015) Multigene knockout utilizing off-target mutations of the CRISPR/Cas9 system in rice. Plant Cell Physiol 56:41–47. doi:10.1093/pcp/pcu154
Fang J, Chai C, Qian Q, Li C, Tang J, Sun L, Huang Z, Guo X, Sun C, Liu M, Zhang Y, Lu Q, Wang Y, Lu C, Han B, Chen F, Cheng Z, Chu C (2008) Mutations of genes in synthesis of the carotenoid precursors of ABA lead to pre-harvest sprouting and photo-oxidation in rice. Plant J 54:177–189. doi:10.1111/j.1365-313X.2008.03411.x
Fauser F, Schiml S, Puchta H (2014) Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J 79:348–359. doi:10.1111/tpj.12554
Feng Z, Zhang B, Ding W, Liu X, Yang DL, Wei P, Cao F, Zhu S, Zhang F, Mao Y, Zhu JK (2013) Efficient genome editing in plants using a CRISPR/Cas system. Cell Res 23:1229–1232. doi:10.1038/cr.2013.114
Feng Z, Mao Y, Xu N, Zhang B, Wei P, Yang DL, Wang Z, Zhang Z, Zheng R, Yang L, Zeng L, Liu X, Zhu JK (2014) Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc Natl Acad Sci 111:4632–4637. doi:10.1073/pnas.1400822111
Gaj T, Gersbach CA, Barbas CF (2013) ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol 31:397–405. doi:10.1016/j.tibtech.2013.04.004
Gupta R, Musunuru K (2014) Expanding the genetic editing tool kit: ZFNs, TALENs, and CRISPR-Cas9. J Clin Invest 124:4154–4161. doi:10.1172/jci72992
Hable WE, Oishi KK, Schumaker KS (1998) Viviparous-5 encodes phytoene desaturase, an enzyme essential for abscisic acid (ABA) accumulation and seed development in maize. Mol Gen Genet 257:167–176. doi:10.1007/s004380050636
Hartung F, Schiemann J (2014) Precise plant breeding using new genome editing techniques: opportunities, safety and regulation in the EU. Plant J 78:742–752. doi:10.1111/tpj.12413
Hiei Y, Komari T (2006) Improved protocols for transformation of indica rice mediated by Agrobacterium tumefaciens. Plant Cell Tiss Organ Cult. doi:10.1007/s11240-005-9069-8
Hiei Y, Komari T (2008) Agrobacterium-mediated transformation of rice using immature embryos or calli induced from mature seed. Nat Protoc 3:824–834. doi:10.1038/nprot.2008.46
Hsu PD, Lander ES, Zhang F (2014) Development and applications of CRISPR-Cas9 for genome engineering. Cell 157:1262–1278. doi:10.1016/j.cell.2014.05.010
Ishizaki T, Kumashiro T (2008) Genetic transformation of NERICA, interspecific hybrid rice between Oryza glaberrima and O. sativa, mediated by Agrobacterium tumefaciens. Plant Cell Rep 27:319–327. doi:10.1007/s00299-007-0465-x
Jiang W, Yang B, Weeks DP (2014) Efficient CRISPR/Cas9-mediated gene editing in Arabidopsis thaliana and inheritance of modified genes in the T2 and T3 generations. PLoS ONE 9:e99225. doi:10.1371/journal.pone.0099225
Joung JK, Sander JD (2013) TALENs: a widely applicable technology for targeted genome editing. Nat Rev Mol Cell Biol 14:49–55. doi:10.1038/nrm3486
Kaneda (2007) Breeding and dissemination efforts of “NERICA” (4) efforts for dissemination of NERICAs in African countries. Jpn J Trop Agr 51:145–151
Kawahara Y, Bastide M, Hamilton JP, Kanamori H, McCombie WR, Ouyang S, Schwartz DC, Tanaka T, Wu J, Zhou S, Childs KL, Davidson RM, Lin H, Quesada-Ocampo L, Vaillancourt B, Sakai H, Lee SS, Kim J, Numa H, Itoh T, Buell CR, Matsumoto T (2013) Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice (N Y) 6:4. doi:10.1186/1939-8433-6-4
Khush GS (1987) Rice breeding: past, present and future. J Genet 66:195–216. doi:10.1007/BF02927713
Lei Y, Lu L, Liu HY, Li S, Xing F, Chen LL (2014) CRISPR-P: a web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol Plant 7:1494–1496
Li T, Liu B, Spalding M, Weeks DP, Yang B (2012) High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat Biotechnol 30:390–392. doi:10.1038/nbt.2199
Mao Y, Zhang H, Xu N, Zhang B, Gou F, Zhu JK (2013) Application of the CRISPR–Cas system for efficient genome engineering in plants. Mol Plant 6:2008–2011. doi:10.1093/mp/sst121
Miao J, Guo D, Zhang J, Huang Q, Qin G, Zhang X, Wan J, Gu H, Q LJ (2013) Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res 23:1233–1236. doi: 10.1038/cr.2013.123
Mikami M, Toki S, Endo M (2015) Parameters affecting frequency of CRISPR/Cas9 mediated targeted mutagenesis in rice. Plant Cell Rep 34:1807–1815. doi:10.1007/s00299-015-1826-5
Mohanty A, Sarma NP, Tyagi A (1999) Agrobacterium-mediated high frequency transformation of an elite indica rice variety Pusa basmati 1 and transmission of the transgenes to R2 progeny. Plant Sci 147:127–137. doi:10.1016/S0168-9452(99)00103-X
Naito Y, Hino K, Bono H, Ui-Tei K (2015) CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics 31:1120–1123. doi:10.1093/bioinformatics/btu743
Nitta Y (2010) Japanese rice producers’ shift from high yield to high palatability and quality—characteristics of highly palatable rice. Journal of Developments in Sustainable Agriculture 5:96–100
Ozawa K (2009) Establishment of a high efficiency Agrobacterium-mediated transformation system of rice (Oryza sativa L.). Plant Sci 176:522–527. doi:10.1016/j.plantsci.2009.01.01
Pan C, Ye L, Qin L, Liu X, He Y, Wang J, Chen L, Lu G (2016) CRISPR/Cas9-mediated efficient and heritable targeted mutagenesis in tomato plants in the first and later generations. Sci Rep 6:24765. doi:10.1038/srep24765
Petolino JF (2015) Genome editing in plants via designed zinc finger nucleases. In Vitro Cell Dev Biol, Plant 51:1–8. doi:10.1007/s11627-015-9663-3
Project I (2005) The map-based sequence of the rice genome. Nature 436:793–800. doi:10.1038/nature03895
Qin G, Gu H, Ma L, Peng Y, Deng XW, Chen Z, Qu LJ (2007) Disruption of phytoene desaturase gene results in albino and dwarf phenotypes in Arabidopsis by impairing chlorophyll, carotenoid, and gibberellin biosynthesis. Cell Res 17:471–482. doi:10.1038/cr.2007.40
Rachmawati D, Hosaka T, Inoue E, Anzai H (2004) Agrobacterium-mediated transformation of Javanica rice cv. Rojolele. Biosci Biotechnol Biochem 68:1193–1200. doi:10.1271/bbb.68.1193
Shan Q, Wang Y, Li J et al (2013) Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotech 31:686–688. doi:10.1038/nbt.2650
Shan Q, Wang Y, Li J, Gao C (2014) Genome editing in rice and wheat using the CRISPR/Cas system. Nat Protoc 9:2395–2410. doi:10.1038/nprot.2014.157
Sorek R, Lawrence CM, Wiedenheft B (2013) CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu Rev Biochem 82:237–266
Xie K, Minkenberg B, Yang Y (2014) Targeted gene mutation in rice using a CRISPR-Cas9 system. Bio Protocol 4:e1225
Xu R, Li H, Qin R, Wang L, Li L, Wei P, Yang J (2014) Gene targeting using the Agrobacterium tumefaciens-mediated CRISPR-Cas system in rice. Rice (N Y) 7:5. doi:10.1186/s12284-014-0005-6
Xu RF, Li H, Qin RY, Li J, Qiu CH, Yang YC, Ma H, Li L, Wei PC, Yang JB (2015) Generation of inheritable and “transgene clean” targeted genome-modified rice in later generations using the CRISPR/Cas9 system. Sci Rep 5:11491. doi:10.1038/srep11491
Zhang F, Maeder ML, Unger-Wallace E, Hoshaw JP, Reyon D, Christian M, Li X, Pierick CJ, Dobbes D, Peterson T, Joung JK, Voytas DF (2010) High frequency targeted mutagenesis in Arabidopsis thaliana using zinc finger nucleases. Proc Natl Acad Sci U S A 107:12028–12033. doi:10.1073/pnas.0914991107
Zhang H, Zhang J, Wei P, Zhang B, Gou F, Feng Z, Mao Y, Yang L, Zhang H, Xu N, Zhu JK (2014) The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotech J 12:797–807. doi:10.1111/pbi.12200
Zhou H, Liu B, Weeks DP, Spalding MH, Yang B (2014) Large chromosomal deletions and heritable small genetic changes induced by CRISPR/Cas9 in rice. Nucleic Acids Res 42:10903–10914. doi:10.1093/nar/gku806
Zhu C, Bortesi L, Baysal C, Twyman RM, Fischer R, Capell T, Schillberg S, Christou P (2016) Characteristics of genome editing mutations in cereal crops. Trends Plant Sci. doi:10.1016/j.tplants.2016.08.009
Acknowledgements
I thank the Africa Rice Center for providing the seeds of NERICA1 and the International Center for Tropical Agriculture for providing the seeds of Curinga. This work was supported by grants from the Ministry of Agriculture, Forestry, and Fisheries of Japan. I am grateful for the excellent technical support and valuable suggestions provided by Dr. Yasunari Fujita, Dr. Yukari Nagatosi, Eiko Tamaki, Ryoko Agata, Uday Mitsuyasu, Emiko Kishi, Megumu Yano, and Masami Toyoshima of the Japan International Research Center for Agricultural Sciences.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The author declares that he has no conflict of interest.
Electronic supplementary material
ESM 1
(PDF 5120 kb)
Rights and permissions
About this article
Cite this article
Ishizaki, T. CRISPR/Cas9 in rice can induce new mutations in later generations, leading to chimerism and unpredicted segregation of the targeted mutation. Mol Breeding 36, 165 (2016). https://doi.org/10.1007/s11032-016-0591-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-016-0591-7