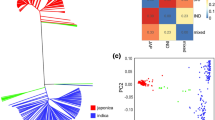
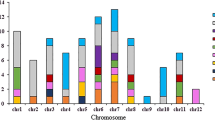
Abstract
Seed vigor is an important seed quality trait in rice (Oryza sativa L.) that profoundly affects seedling morphogenesis in different environments. Generally, highly vigorous seeds have many advantages in agricultural production, such as resistance to adverse stresses, rapid emergence, and yield improvement. In the present study, one germplasm collection and one recombinant inbred line (RIL) population were used to identify quantitative trait loci (QTLs) for seed vigor. A total of 19 single nucleotide polymorphisms (SNPs) were found to be significantly associated with seed vigor (−log10(P) > 6) in the germplasm collection, which consisted of 200 rice cultivars. Nine loci responsible for seed vigor were identified via QTL mapping by using a high-density bin map. To screen the candidate genes more efficiently, we selected six loci that were co-localized in GWAS and QTL mapping, overlapped previous reports, repeat detected across two seasons, or high contribution rate as reliable loci. A total of 44 differentially expressed genes were obtained from the reliable loci via gene expression profile analysis. Among these 44 genes, Os06g0108600, Os06g0110200, Os06g0253100, Os06g0282000, Os07g0583600, Os07g0592600, and Os09g0432300 were the most promising candidates associated with seed vigor.







Similar content being viewed by others
Availability of data and material
All data generated or analyzed during this study are included in this published article.
Abbreviations
- GR:
-
germination rate
- GP:
-
germination potential
- GI:
-
germination index
- VI:
-
vigor index
- QTL:
-
quantitative trait loci
- GWAS:
-
genome-wide association study
- DSR:
-
direct-seeded rice
- WS:
-
wet season
- DS:
-
dry season
References
Ahrazem O, Rubio-Moraga A, Trapero-Mozos A, Lopez Climent MF, Gomez-Cadenas A, Gomez-Gomez L (2015) Ectopic expression of a stress-inducible glycosyltransferase from saffron enhances salt and oxidative stress tolerance in Arabidopsis while alters anchor root formation. Plant Sci 234:60–73
Bewley JD, Bradford KJ, Hilhorst H (2012). Seeds: physiology of development, germination and dormancy. Springer Science & Business Media
Chen F, Bradford KJ (2000) Expression of an expansin is associated with endosperm weakening during tomato seed germination. Plant Physiol 124(3):1265–1274
Cheng J, He Y, Yang B, Lai Y, Wang Z, Zhang H (2015) Association mapping of seed germination and seedling growth at three conditions in indica rice (Oryza sativa L.). Euphytica 206(1):103–115
Chen L, Gao W, Chen S, Wang L, Zou J, Liu Y, Wang H, Chen Z, Guo T (2016) High-resolution QTL mapping for grain appearance traits and co-localization of chalkiness-associated differentially expressed candidate genes in rice. Rice 9:1–17
Dong T, Hwang I (2014) Contribution of ABA UDP-glucosyltransferases in coordination of ABA biosynthesis and catabolism for ABA homeostasis. Plant Signal Behav 9:e28888
Ellis RH (1992) Seed and seedling vigour in relation to crop growth and yield. Plant Growth Regul 11(3):249–255
Foolad MR, Subbiah P, Zhang L (2007) Common QTL affect the rate of tomato seed germination under different stress and nonstress conditions. Int J Plant Sci 2007:1–10
Fujino K, Sekiguchi H, Matsuda Y, Sugimoto K, Ono K, Yano M (2008) Molecular identification of a major quantitative trait locus, qLTG3-1, controlling low-temperature germinability in rice. P Natl Acad Sci USA 105(34):12623–12628
Hayashi E, Aoyama N, Still DW (2008) Quantitative trait loci associated with lettuce seed germination under different temperature and light environments. Genome 51(11):928–947
Howell KA, Narsai R, Carroll A, Ivanova A, Lohse M, Usadel B, Millar AH, Whelan J (2008) Mapping metabolic and transcript temporal switches during germination in rice highlights specific transcription factors and the role of RNA instability in the germination process. Plant Physiol 149(2):961–980
Hsu S, Tung C (2015) Genetic mapping of anaerobic germination-associated QTLs controlling coleoptile elongation in rice. Rice 8:38
Huang J, Zhang J, Li W, Hu W, Duan L, Feng Y, Qiu F, Yue B (2013) Genome-wide association analysis of ten chilling tolerance indices at the germination and seedling stages in maize. J Integr Plant Biol 55(8):735–744
Jung J, Park C (2011) Auxin modulation of salt stress signaling in Arabidopsis seed germination. Plant Signal Behav 6:1198–1200
Lander ES, Schork NJ (1994) Genetic dissection of complex traits. Science 265:2037–2048
Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet J, Subramanian A, Ross KN, Reich M, Hieronymus H, Wei G, Armstrong SA, Haggarty SJ, Clemons PA, Wei R, Carr SA, Lander ES, Golub TR (2006) The connectivity map: using gene-expression signatures to connect small molecules. Genes Dis Sci 313:1929–1935
Li CC (1969) Population subdivision with respect to multiple alleles. Ann Hum Genet 33(1):23–29
Li Y, Huang Y, Bergelson J, Nordborg M, Borevitz JO (2010) Association mapping of local climate-sensitive quantitative trait loci in Arabidopsis thaliana. P Natl Acad Sci USA 107(49):21199–21204
Li J, Xu R, Wang C, Qi L, Zheng X, Wang W, Ding Y, Zhang L, Wang Y, Cheng Y, Zhang L, Qiao W, Yang Q (2018) A heading date QTL, qHD7.2, from wild rice (Oryza rufipogon) delays flowering and shortens panicle length under long-day conditions. Sci Rep-Uk 8:2928
Liu L, Lai Y, Cheng J, Wang L, Du W, Wang Z, Zhang H (2014) Dynamic quantitative trait locus analysis of seed vigor at three maturity stages in rice. PLoS One 9(12):e115732
Lu Y, Zhang S, Shah T, Xie C, Hao Z, Li X, Farkhari M, Ribaut JM, Cao M, Rong T, Xu Y (2010) Joint linkage–linkage disequilibrium mapping is a powerful approach to detecting quantitative trait loci underlying drought tolerance in maize. P Natl Acad Sci USA 107(45):19585–19590
Lu Q, Niu X, Zhang M, Wang C, Xu Q, Feng Y, Yang Y, Wang S, Yuan X, Yu H, Wang Y, Chen X, Liang X, Wei X (2018) Genome-wide association study of seed dormancy and the genomic consequences of improvement footprints in Rice (Oryza sativa L.). Front Plant Sci 8:2213
Ma W, Guan X, Li J, Pan R, Wang L, Liu F, Ma H, Zhu S, Hu J, Ruan YL, Chen X, Zhang T (2019) Mitochondrial small heat shock protein mediates seed germination via thermal sensing. P Natl Acad Sci USA 116:4716–4721. https://doi.org/10.1073/pnas.1815790116
Magwa RA, Zhao H, Xing Y (2016) Genome-wide association mapping revealed a diverse genetic basis of seed dormancy across subpopulations in rice (Oryza sativa L.). BMC Genet 17:28
Meng L, Li H, Zhang L, Wang J (2015) QTL IciMapping: integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J 3:269–283
Narsai R, Howell KA, Carroll A, Ivanova A, Millar AH, Whelan J (2009) Defining core metabolic and transcriptomic responses to oxygen availability in rice embryos and young seedlings. Plant Physiol 151:306–322
Park J, Kim YS, Kim SG, Jung JH, Woo JC, Park CM (2011) Integration of auxin and salt signals by the NAC transcription factor NTM2 during seed germination in Arabidopsis. Plant Physiol 156(2):537–549
Poland JA, Brown PJ, Sorrells ME, Jannink JL (2012) Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PLoS One 7(2):e32253
Rajjou L, Duval M, Gallardo K, Catusse J, Bally J, Job C, Job D (2012) Seed germination and vigor. Annu Rev Plant Biol 63:507–533
Rafalski JA (2010) Association genetics in crop improvement. Curr Opin Plant Biol 13(2):174–180
Robinson MD, McCarthy DJ, Smyth GK (2009) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26(1):139–140
Sasaki T, Burr B (2000) International Rice Genome Sequencing Project: the effort to completely sequence the rice genome. Curr Opin Plant Biol 3(2):138–142
Sales E, Viruel J, Domingo C, Marqués L (2017) Genome wide association analysis of cold tolerance at germination in temperate japonica rice (Oryza sativa L.) varieties. PLoS One 12(8):e183416
Sánchez-Montesino R, Bouza-Morcillo L, Marquez J, Ghita M, Duran-Nebreda S, Gomez L, Holdsworth MJ, Bassel G, Onate-Sanchez L (2019) A regulatory module controlling GA-mediated endosperm cell expansion is critical for seed germination in Arabidopsis. Mol Plant 12(1):71–85
Schläppi MR, Jackson AK, Eizenga GC, Wang A, Chu C, Shi Y, Shimoyama N, Boykin DL (2017) Assessment of five chilling tolerance traits and GWAS mapping in rice using the USDA mini-Core collection. Front Plant Sci 8:957
Sechet J, Frey A, Cuzzi D, Berger A, Perreau F, Cueff G, Charif D, Rajjou L, Mouille G, North HM (2016) Xyloglucan metabolism differentially impacts the cell wall characteristics of the endosperm and embryo during Arabidopsis seed germination. Plant Physiol. Marion-Poll A 170:1367–1380
Sonah H, Bastien M, Iquira E, Tardivel A, Legare G, Boyle B, Normandeau E, Laroche J, Larose S, Jean M, Belzile F (2013) An improved genotyping by sequencing (GBS) approach offering increased versatility and efficiency of SNP discovery and genotyping. PLoS One 8(1):e54603
Soós V, Juhász A, Light ME, Van Staden J, Balázs E (2009) Smoke-water-induced changes of expression pattern in Grand Rapids lettuce achenes. Seed Sci Res 19(1):37–49
Sun Q, Wang JH, Sun BQ (2007) Advances on seed vigor physiological and genetic mechanisms. Agr Sci CHINA 6(9):1060–1066
Tao Y, Jiang L, Liu Q, Zhang Y, Zhang R, Ingvardsen CR, Frei UK, Wang B, Lai J, Lübberstedt T, Xu M (2013) Combined linkage and association mapping reveals candidates for Scmv1, a major locus involved in resistance to sugarcane mosaic virus (SCMV) in maize. BMC Plant Biol 13:162
Tian F, Bradbury PJ, Brown PJ, Hung H, Sun Q, Flint-Garcia S, Rocheford TR, McMullen MD, Holland JB, Buckler ES (2011) Genome-wide association study of leaf architecture in the maize nested association mapping population. Nat Genet 43:159–162
Voegele A, Linkies A, Muller K, Leubner-Metzger G (2011) Members of the GIBBERELLIN receptor gene family GID1 (GIBBERELLIN INSENSITIVE DWARF1) play distinct roles during Lepidium sativum and Arabidopsis thaliana seed germination. J Exp Bot 62(14):5131–5147
Wang ZF, Wang JF, Bao YM, Wang FH, Zhang HS (2010) Quantitative trait loci analysis for rice seed vigor during the germination stage. J ZheJiang Univ-Sc B 11(12):958–964
Wang X, Yu K, Li H, Peng Q, Chen F, Zhang W, Chen S, Hu M, Zhang J (2015) High-density SNP map construction and QTL identification for the apetalous character in Brassica napus L. Front Plant Sci 6:1164
Wang X, Zou B, Shao Q, Cui Y, Lu S, Zhang Y, Huang Q, Huang J, Hua J (2018) Natural variation reveals that OsSAP16 controls low-temperature germination in rice. J Exp Bot 69(3):413–421
Xiao B, Huang Y, Tang N, Xiong L (2007) Over-expression of a LEA gene in rice improves drought resistance under the field conditions. Theor Appl Genet 115(1):35–46
Xie W, Feng Q, Yu H, Huang X, Zhao Q, Xing Y, Yu S, Han B, Zhang Q (2010) Parent-independent genotyping for constructing an ultrahigh-density linkage map based on population sequencing. P Natl Acad Sci USA 107(23):10578–10583
Yu H, Xie W, Wang J, Xing Y, Xu C, Li X, Xiao J, Zhang Q (2011) Gains in QTL detection using an ultra-high density SNP map based on population sequencing relative to traditional RFLP/SSR markers. PLoS One 6(3):e17595
Zhang M, Lu Q, Wu W, Niu X, Wang C, Feng Y, Xu Q, Wang S, Yuan X, Yu H, Wang Y, Wei X (2017) Association mapping reveals novel genetic loci contributing to flooding tolerance during germination in indica rice. Front Plant Sci 8:678
Zhao G, Zhong T (2013) Influence of exogenous IAA and GA on seed germination, vigor and their endogenous levels in Cunninghamia lanceolata. Scand J Forest Res 28(6):511–517
Zhou YL (2011) Function analysis of Nelumbo nucifera metallothionein and heat shock protein genes in seed vigor. Dissertation, Sun Yat-sen University
Acknowledgments
We thank Novogene Co., Ltd., for assisting in sequencing and bioinformatics analyses. We thank the assistant scientist Berlaine Quime from International Rice Research Institute for the critical reading and modification of the manuscript. We also thank Ling Su and Meng Yang for their suggestions and help for experiments and data analyses.
Funding
Financial support for this research was provided in part by a grant from the Breeding New Varieties of Rice Suitable for Light and Simple Cultivation and Mechanized Production (no. 2017YFD0100104), the National Key Technology Research and Development Program of China (no. 2016YFD0102102), the Science and Technology Project of Guangdong Province (no. 2015B020231011), and the earmarked fund for Modern Agro-Industry Technology Research System (no. CARS-01-12).
Author information
Authors and Affiliations
Contributions
Z.C. and H.W. designed the project and J.Y. performed all the experiments and T.G. wrote the manuscript. D.L., K.S., L.L., W.X., J.W., Y.L., and S.W. assisted in conducting experiments and data analysis. H.W. and Z.C. provided the direction for the study and the correction of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Author details
National Engineering Research Center of Plant Space Breeding, South China Agricultural University, Guangzhou 510642, China.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Fig. S1
Differentially expressed genes between parents at different periods. (PNG 84 kb)
Table S1
Primers used for qRT-PCR. (XLSX 9 kb)
Table S2
Phenotypic data of all germplasms. (XLSX 49 kb)
Table S3
Annotated function of 44 differentially expressed genes identified from the six reliable loci. (XLSX 15 kb)
Rights and permissions
About this article
Cite this article
Guo, T., Yang, J., Li, D. et al. Integrating GWAS, QTL, mapping and RNA-seq to identify candidate genes for seed vigor in rice (Oryza sativa L.). Mol Breeding 39, 87 (2019). https://doi.org/10.1007/s11032-019-0993-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11032-019-0993-4