Abstract
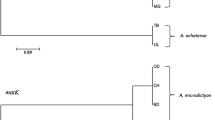
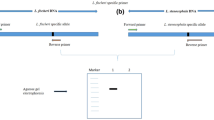
Chrysanthemum indicum L. is a traditional oriental medicinal herb prepared as a tea from flowers that have been used in China and South Korea since ancient times. It has a long history in the treatment of hypertension, inflammation, and respiratory diseases. Among Chrysanthemum species, C. indicum has more active chemical components as well as better therapeutic effects, and C. indicum is mostly used for medicinal purposes in South Korea. However, the usage of C. indicum has become problematic over the years due to the abundance of adulterated Chrysanthemum and confusion with morphologically related species such as C. morifolium, C. boreale, and Aster spathulifolius. Thus, here we developed a method for molecular authentication using chloroplast universal region rpoC2 and morphological authentication based on T-shaped trichomes of the adaxial leaf surface. By using a species-specific primer derived from the rpoC2 region, we established a multiplex allele-specific PCR for the discrimination of C. indicum. Amplicons of 675 bp for C. indicum and 1026 bp for other Chrysanthemum species were produced using both rpoC2-specific and common primers. These primers can be used to analyze dried samples of Chrysanthemum. Morphological discrimination was performed using T-shaped trichomes present only on the adaxial leaf surface of C. indicum species, and then molecular markers were utilized to authenticate C. indicum products from adulterant samples available in the market. Our results indicate that these molecular markers in combination with morphological differentiation can serve as an effective tool for identifying C. indicum.







Similar content being viewed by others
References
Ganie SH, Upadhyay P, Das S, Prasad Sharma M (2015) Authentication of medicinal plants by DNA markers. Plant Gene 4:83–99. https://doi.org/10.1016/j.plgene.2015.10.002
Wang F, Zhang FJ, Di CF et al (2014) Identification of Chrysanthemum (Chrysanthemum morifolium) self-incompatibility. Sci World J. https://doi.org/10.1155/2014/625658
Mitiouchkina TY, Dolgov SV (1998) Modification of Chrysanthemum plant and flower architecture by rolC gene from Agrobacterium rhizogenes introduction. Acta Hortic 508:163–172
Cheng W, Li J, You T, Hu C (2005) Anti-inflammatory and immunomodulatory activities of the extracts from the inflorescence of Chrysanthemum indicum Linné. J Ethnopharmacol 101:334–337. https://doi.org/10.1016/j.jep.2005.04.035
Jeong SC, Kim SM, Jeong YT, Song CH (2013) Hepatoprotective effect of water extract from Chrysanthemum indicum L. flower. Chinese Med (United Kingdom) 8:1–8. https://doi.org/10.1186/1749-8546-8-7
Luyen BTT, Tai BH, Thao NP et al (2015) Anti-inflammatory components of Chrysanthemum indicum flowers. Bioorganic Med Chem Lett 25:266–269. https://doi.org/10.1016/j.bmcl.2014.11.054
Shunying Z, Yang Y, Huaidong Y et al (2005) Chemical composition and antimicrobial activity of the essential oils of Chrysanthemum indicum. J Ethnopharmacol 96:151–158. https://doi.org/10.1016/j.jep.2004.08.031
Chang K-M, Choi E-M, Kim G-H (2010) Chemical constituents of Chrysanthemum indicum L. flower oil and effect on osteoblastic MC3T3-E1 cells. Food Sci Biotechnol 19:815–819
Chang K-M, Kim G-H (2008) Volatile aroma composition of Chrysanthemum indicum L. flower oil. J Food Sci Nutr 13:122–127
Choi H-S, Kim G-H (2011) Volatile flavor composition of gamguk (Chrysanthemum indicum) flower essential oils. Food Sci Biotechnol 20:319–325
Tsuji-Naito K, Saeki H, Hamano M (2009) Inhibitory effects of Chrysanthemum species extracts on formation of advanced glycation end products. Food Chem 116:854–859
Dmitruk M, Weryszko-Chmielewska E (2010) Morphological differentiation and distribution of non-glandular and glandular trichomes on Dracocephalum moldavicum L. shoots. Acta Agrobot 63(1):11–22
Meo AA, Khan MA (2006) Pollen morphology as an aid to the identification of Chrysanthemum species (Compositae-Anthemideae) from Pakistan. Pakistan J Bot 38:29
Sumitomo K, Nishijima T, Onozaki T, Shibata M (2006) Density, length and development of non-glandular trichome on the leaves of wild chrysanthemums and Chrysanthemum cultivars. Hortic Res 5(4):351–356
Bhatt A, Naidoo Y, Nicholas A (2010) An investigation of the glandular and non-glandular foliar trichomes of Orthosiphon labiatus NE Br.[Lamiaceae]. New Zeal J Bot 48:153–161
Dalin P, Ågren J, Björkman C et al (2008) Leaf trichome formation and plant resistance to herbivory. In: Schaller A (ed) Induced plant resistance to herbivory. Springer, Dordrecht, pp 89–105
He J, Chen F, Chen S et al (2011) Chrysanthemum leaf epidermal surface morphology and antioxidant and defense enzyme activity in response to aphid infestation. J Plant Physiol 168:687–693
Buitenhuis R, Shipp L, Scott-Dupree C et al (2014) Host plant effects on the behaviour and performance of Amblyseius swirskii (Acari: Phytoseiidae). Exp Appl Acarol 62:171–180. https://doi.org/10.1007/s10493-013-9735-1
Mircea CC, Cioancă O, Draghia L, Hăncianu M (2015) Morphological characteristics, phenolic and terpenoid profiles in garden Chrysanthemum grown in different nutritional conditions. Not Bot Horti Agrobot Cluj-Napoca 43:371–379
Seethapathy GS, Raclariu-Manolica AC, Anmarkrud JA et al (2019) DNA metabarcoding authentication of ayurvedic herbal products on the european market raises concerns of quality and fidelity. Front Plant Sci 10:1–11. https://doi.org/10.3389/fpls.2019.00068
Kim WJ, Moon BC, Yang S et al (2016) Rapid authentication of the herbal medicine plant species Aralia continentalis Kitag. and Angelica biserrata C.Q. Yuan and R.H. Shan using ITS2 sequences and multiplex-SCAR markers. Molecules. https://doi.org/10.3390/molecules21030270
Chen X, Liao B, Song J et al (2013) A fast SNP identification and analysis of intraspecific variation in the medicinal Panax species based on DNA barcoding. Gene 530:39–43. https://doi.org/10.1016/j.gene.2013.07.097
Lee OR, Kim M-K, Yang D-C (2012) Authentication of medicinal plants by SNP-based multiplex PCR. In: Sucher N, Hennell J, Carles M (eds) Plant DNA Fingerprinting and Barcoding. Methods in molecular biology (Methods and Protocols), vol 862. Humana Press, New York, pp 135–147
Wang H, Kim MK, Kim YJ et al (2012) Molecular authentication of the Oriental medicines Pericarpium Citri Reticulatae and Citri Unshius Pericarpium using SNP markers. Gene 494:92–95. https://doi.org/10.1016/j.gene.2011.11.026
Serino G, Maliga P (1998) RNA polymerase subunits encoded by the plastid rpo genes are not shared with the nucleus-encoded plastid enzyme. Plant Physiol 117:1165–1170. https://doi.org/10.1104/pp.117.4.1165
Corpet F (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 16(22):10881–10890
Hall TA (2011) A User-Friendly Biological Sequence Alignment Editor and Analysis Program for Windows 95/98/NT. In: Nucleic Acids Symp. Ser. https://www.google.com/search?q=bio+edit+software+citation&rlz=1C1RUCY_enKR789KR789&oq=bio+edit+software+citat&aqs=chrome.1.69i57j33.13979j0j7&sourceid=chrome&ie=UTF-8.
Elvin MA, Kelley RB, Drew BT (2019) Trichome morphology relates to taxonomic diversity in Monardella (Lamiaceae) in the lower Snake River Watershed of Idaho and Oregon, USA: taxonomic studies in Monardella (Lamiaceae) VI. J Torrey Bot Soc. https://doi.org/10.3159/TORREY-D-18-00059.1
Riddick EW, Simmons AM (2014) Do plant trichomes cause more harm than good to predatory insects? Pest Manag Sci 70:1655–1665
Stavrinides MC, Skirvin DJ (2003) The effect of chrysanthemum leaf trichome density and prey spatial distribution on predation of Tetranychus urticae (Acari: Tetranychidae) by Phytoseiulus persimilis (Acari: Phytoseiidae). Bull Entomol Res 93:343–350. https://doi.org/10.1079/ber2003243
Abid S, Mohanan P, Kaliraj L et al (2019) Development of species-specific chloroplast markers for the authentication of Gynostemma pentaphyllum and their distribution in the Korean peninsula. Fitoterapia 138:104295. https://doi.org/10.1016/j.fitote.2019.104295
Kim SJ, Lee CH, Kim J, Kim KS (2014) Phylogenetic analysis of Korean native Chrysanthemum species based on morphological characteristics. Sci Hortic (Amsterdam) 175:278–289. https://doi.org/10.1016/j.scienta.2014.06.018
Klie M, Schie S, Linde M, Debener T (2014) The type of ploidy of chrysanthemum is not black or white: a comparison of a molecular approach to published cytological methods. Front Plant Sci 5:1–8. https://doi.org/10.3389/fpls.2014.00479
Zhang Y, Wang C, Ma HZ, Dai SL (2013) Assessing the genetic diversity of Chrysanthemum cultivars with microsatellites. J Am Soc Hortic Sci 138:479–486. https://doi.org/10.21273/jashs.138.6.479
Mohanan P, Hurh J, Kim SO et al (2019) Chloroplast DNA-derived markers for the authentication of oriental medicinal Rubus species and mistaken identity of bokbunja in the local markets of Korea. Plant Biotechnol Rep 13:305–314. https://doi.org/10.1007/s11816-019-00540-5
Fang W, Meinhardt LW, Tan H et al (2016) Identification of the varietal origin of processed loose-leaf tea based on analysis of a single leaf by SNP nanofluidic array. Crop J. https://doi.org/10.1016/j.cj.2016.02.001
Funding
This study was supported by a grant from the Korea Institute of Planning & Evaluation for Technology in Food, Agriculture, Forestry & Fisheries (KIPET NO: 119111-01), Republic of Korea.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflicts of interest
The authors declare that they have no conflicts of interest.
Ethical approval
This chapter does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Abid, S., Kaliraj, L., Arif, M.H. et al. Molecular and morphological discrimination of Chrysanthemum indicum using allele-specific PCR and T-shaped trichome. Mol Biol Rep 47, 7699–7708 (2020). https://doi.org/10.1007/s11033-020-05844-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-020-05844-2