Abstract
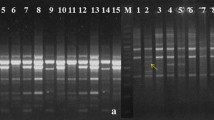
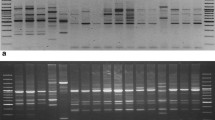
Gloriosa superba L., an endangered medicinal plant with global interest due to presence of colchicine, an important alkaloid used in formulations of Indian and Traditional medicine. The plant has become endangered due to its unscientifically exploitation and high medicinal values. In the Present study 10 randomly amplified polymorphic DNA (RAPD) and 6 ISSR markers were employed to assess genetic divergence among micro propagated, wild and field cultivated plants of Gloriosa superba collected from different parts of India. In RAPD analysis, all the 10 accession with 10 RAPD primers amplified 466 fragments, with 96.43 % polymorphism and with an average of 46.6 bands per primer. The size of amplicons varied from 1656 to 100 bp. While, ISSR primers produced 328 fragments of which 298 were polymorphic with an average of 49.7 bands per primer with 91.83% polymorphism. The size of amplicons ranges from 2395 to 181 bp. RAPD, ISSR markers were also assessed by calculating polymorphic information content (PIC) to discriminate the genotypes, Average PIC value for RAPD, ISSR and combined RAPD + ISSR markers obtained was ≤ 0.50 suggesting the informativeness of markers. Jaccard’s coefficient ranges from 0.18 to 0.75 (RAPD) and 0.17 to 0.61 (ISSR) and 0.21–0.52 for pooled ISSR and RAPD markers. The clustering pattern based on UPGMA analysis of the genotypes in the combined analysis revealed that the majority of the genotypes remained similar to the ISSR dendrogram, while the RAPD-based dendrogram showed some variation in the clustering of genotypes. The result of PCA scattered plot obtained were in agreement with the UPGMA dendrogram, which further confirms the genetic relationships explain by cluster analysis. Results confirmed that the genotype studied had good genetic diversity and can be used for identification, conservation, and future breeding program of Gloriosa species and consequently for the benefit of the pharmaceutical industries.







Similar content being viewed by others
Abbreviations
- RAPD:
-
Random amplified polymorphic DNA
- ISSR:
-
Inter simple sequence repeats
- RFLP:
-
Restriction fragment length polymorphism
- AFLP:
-
Amplified fragment length polymorphism
- PIC:
-
Polymorphic information content
- DNA:
-
Deoxyribonucleic acid
- PCR:
-
Polymerase chain reaction
- UPGMA:
-
Unweighted pair group method with the arithmetic averaging algorithm
- PCA:
-
Principal component analysis
References
Jomy J, Jennifer F, Nandgude Tanaji D, Niphade S, Salva A, Deshmukh PT (2009) Analgesic and anti-inflammatory activities of the hydroalcoholic extract from Gloriosa superba Linn. Int J Green Phar. https://doi.org/10.4103/0973-8258.56277
Hemaiswarya S, Raja R, Anbazhagan C, Venkatesan V (2009) Antimicrobial and mutagenic properties of the root tubers of Gloriosa superba Linn (Kalihari). Pakistan J Bot 41(1):293–299
Kee NLA, Mnonopi N, Davids H, Naude RJ, Frost CL (2008) Antithrombotic/anticoagulant and anticancer activities of selected medicinal plants from South Africa. African J Biotechnol 7(3):217–223
Haroon K, Murad AK, Iqbal H (2008) Enzyme inhibition activities of the extracts from rhizomes of Gloriosa superba Linn (Colchicaceae). J Enzyme Inhibit Med Chem 22(6):722–725
Reuter S, Prasad S, Phromnoi K, Ravindran J, Sung B, Yadav RV, Kannappan R (2010) Rhizomes of Gloriosa superba Linn (Colchicaceae). J Enzyme Inhibit Med Chem 22(6):722–725
Rokade SS, Joshi KA, Mahajan K, Patil S, Tomar G, Dubal DS, Parihar VS, Kitture R, Bellare JR, Ghosh S (2018) Gloriosa superba mediated synthesis of platinum and palladium nanoparticles for induction of apoptosis in breast cancer. Bioinorg Chem Appl. https://doi.org/10.1155/2018/4924186
Balkrishna A, Das SK, Pokhrel S, Joshi A, Laxmi VS, Sharma VK, Sharma V, Sharma N, Joshi CS (2019) Colchicine: isolation, LC–MS QTof screening, and anticancer activity study of Gloriosa superba seeds. Molecules. https://doi.org/10.3390/molecules24152772
Mohandass S, Indhumathi T (2011) Hepatoprotective efficacy of Gloriosa superba Linn. against 25. paracetamol treated experimental rats-An in vivo study. Golden Res Thoughts 1(4):1–4
Jothi U, Jebamalar JA, Sivakumar T (2019) Study on estimation and antioxidant activity of gloriosa superba l whole plant extract. Int J Sci Res Biol Sci 6(3):50–55
Pawar BM, Wavhal VP, Pawar ND, Agarwal MR, Shinde PB, Kamble HV (2010) Anthelmintic activity of Gloriosa superba Linn (Liliaceae). Int J PharmTech Res 205:1483–1487
Riaz MNA, Chennamaneni SR, Challa SR (2013) Anti-ulcerogenic evaluation of Gloriosa Superba tuber extracts in-vivo and anthelmintic activity in-vitro: a comparison. J Med Health Sci 24:73–79
Nautiyal OP (2011) Isolation of 3-demethylcolchicine from Gloriosa superba sludge and coupling with α- acetobromoglucose to yield colchicoside and thiocolchicoside. J Nat Prod 4:87–93
Kuo MC, Chang SJ, Hsieh MC (2015) Colchicine Significantly Reduces Incident Cancer in Gout Male Patients A 12-Year Cohort Study. Medicine 94:1–6
Sharma S, Sharma YP, Thakur P (2017) Quantification of colchicine in different parts of Gloriosa superba L. IJCS 53:147–149
Sivakumar T, Gajalakshmi D (2019) Phyto chemical analysis and evaluation of antimicrobial activity in the whole plant extract of Gloriosa superba. Asian J Pharmac Clin Res 126:245–249. https://doi.org/10.22159/ajpcr.2019.v12i6.33059
Muthukrishnan SD, Subramaniyan A (2012) Phytochemical constituents of Gloriosa superba seed, tuber and leaves. Res J Pharmac Biol Chem Sci 33:111–117
Pattanaik C, Reddy CS, Reddy KN (2009) Ethno-medicinal survey of threatened plants in Eastern Ghats. India Our Nature 7:122–128
Mishra SB, Dwivedi S, Shashi A, Prajapati K (2008) Ethnomedicinal uses of some plant species by ethnic and rural peoples of the salem district of tamil nadu with special reference to the conservation of vanishing species. Ethnobot Leaflets 12:873–87
Mahajan R (2015) Gloriosa superba L.: an Endangered Medicinal Plant. Hort Flora Res Spectrum 42:168–171
Ranjith KR, Rohini A (2019) Economic analysis of production and marketing of Gloriosa superba L Tamil Nadu. IJCS 7(3):4275–4278
Nag A, Ahuja PS, Sharma RK (2015) Genetic diversity of high-elevation populations of an endangered medicinal plant. AoB PLANTS. https://doi.org/10.1093/aobpla/plu076
Grover A, Sharma PC (2016) Development and use of molecular markers: past and present. Crit Rev Biotechnol 362:290–302
Petit RJ, Hampe A (2006) Some evolutionary consequences of being a tree. Annu Rev Ecol Evol Syst 37:187–214. https://doi.org/10.1146/annurev.ecolsys.37.091305.110215
Prajapat P, Sasidharan N, Ballani A (2015) Assessment of genetic diversity in four brassica species using randomly amplified polymorphic DNA markers. Int J Agric Environ Biotechnol 84:831–836
Bhattacharyya P, Kumaria S (2015) Molecular characterization of Dendrobium nobile Lindl., an endangered medicinal orchid, based on randomly amplified polymorphic DNA. Plant Syst Evol 3011:201–210
Salazar-Laureles ME, Pérez López DDJ, González- Huerta A, Vázquez-García LM, Valadez-Moctezuma E (2015) Genetic variability analysis of faba bean accessions using Inter-simple sequence repeat (ISSR) markers. Chilean J Agric Res. https://doi.org/10.4067/S0718-58392015000100017
Asfaw BM, Dagne K, Keneni G, Kemal S, Kassahu T (2018) Genetic diversity study of Ethiopian Faba bean (Vicia faba L.) varieties based on phenotypic traits and inter simple sequence repeat (ISSR) markers. African J Biotechnol. https://doi.org/10.5897/AJB2017.16331
Rajagopal C, Kandhasamy R (2013) Assessment of genetic diversity of Gloriosa superba L. accessions detected by random amplified polymorphic DNA analysis. J Med Plants Res 7(28):2122–2127. https://doi.org/10.5897/JMPR12.805
Yadav K, Aggarwal A, Singh N (2013) Evaluation of genetic fidelity among micropropagated plants of Gloriosa superba L using DNA-based markers— a potential medicinal plant. Fitoterapia 89:265–270. https://doi.org/10.1016/j.fitote.2013.06.009
Paul Jasmine JA, Balakrishnan V (2018) Intra S pecific Analysis of Gloriosa superba (L.) through ISSR finger printing and DNA sequencing of ecotypes collected from different accessions of Tamil Nadu State, India. Res Plant Biol 8:17–21. https://doi.org/10.25081/ripb.2018.v8.3600
Sahana KS, Gnanam R, Rajesh S, Rajamani K (2019) Evaluation of genetic diversity in Gloriosa superba L, an endangered medicinal plant using molecular marker. Int J Curr Microbiol App Sci 86:2125–2134
Patel HK, Fougat RS, Kumar S, Mistry JG, Kumar M (2015) Detection of genetic variation in Ocimum species using RAPD and ISSR markers. 3 Biotech 5(5):697–707. https://doi.org/10.1007/s13205-014-0269-y
Gupta M, Chyi Y-S, Romero-Severson J, Owen JL (1994) Amplification of DNA markers from evolutionarily diverse gen-omes using single primers of simple-sequence repeats. TheorAppl Genet 89:998–1006
Godwin ID, Aitken EA, Smith LW (1997) Application of inter simple sequence repeat ISSR markers to plant genetics. Electrophoresis 189:1524–1528
Virk PS, Zhu J, John Newbury H, Bryan G, Jackson MT, Ford-Lloyd BV (2012) Effectiveness of different classes of molecular marker for classifying and revealing variation in rice (Oryza sativa) germplasm. Euphytica 112(3):275–284. https://doi.org/10.1023/A:1003952720758
Tiwari JK, Chandel P, Gupta S, Gopal J, Singh BP, Bhardwaj V (2013) Analysis of genetic stability of in vitro propagated potato microtubers using DNA markers. Physiol Mol Biol Plants: Int J Funct Plant Biol 194:587–595. https://doi.org/10.1007/s12298-013-0190-6
Badar Z et al (2017) Analysis of genetic fidelity of wild type and in vitro regenerated aloe vera plants through RAPD and ISSR molecular markers. Int J Biotech Bioeng 3(8):259–267
Khatik N, Joshi R (2020) Assessment of genetic fidelity in microclones of curry leaf plants [Murraya koenigii L. Spreng.] using ISSR markers. Indian J Exp Biol 584:286–291
Shasany AK, Darokar MP, Sakia D, Rajkumar S, Sundaresan V, Khanuja SPS (2003) Genetic diversity and species relationship in Asparagus spp. using RAPD analysis. J Med Arom Plant Sci 25:698–704
Singh R and Randhawa G (2008) Comparative assessment of genetic diversity in Indian and exotic neem (Azadirachta indica) using AFLP markers Indian J Agricul Sci 78(10)
Tripathi N, Saini N, Tiwari S (2011) Assessment of genetic diversity among Aloe vera accessions using amplified fragment length polymorphism. Int J Med Arom Plants 1:115–121
Tilwari A, Tamrakar K, Sharma R (2013) Use of random amplified polymorphic DNA (RAPD) for assessing genetic diversity of Ocimum sanctum (Krishna Tulsi) from different environments of Central India. J Med Plant Res. https://doi.org/10.5897/JMPR12.429
Tilwari A, Chauhan D, Sharma R, Singh RK (2016) Assessment of genetic variations among medicinal plant cassia tora from different geographic regions of central india using RAPD markers medicinal and aromatic plants. Med Aromat Plants (Los Angel) 5:6. https://doi.org/10.4172/2167-0412.1000276
Sharma D, Khatik N, Joshi R (2019) Evaluation of genetic stability of Micropropagated plnts of Murraya Koenigii L. Spreng. Using RAPD Markers. Asian J Microbiol Biotechnol Environ Sci 211:146–151
Akçali Giachino RR (2020) Investigation of the genetic variation of anise Pimpinella anisum L. using RAPD and ISSR markers. Genet Resour Crop Evol 67:763–780. https://doi.org/10.1007/s10722-019-00861-y
Samira A, Osman, Honda BM, Ali (2020) Genetic diversity of five lathyrus species using rapd, issr and scot markers. Asian J Plant Sci 19:152–165
Gogoi B, Wann SB, Saikia SP (2020) Comparative assessment of ISSR, RAPD, and SCoT markers for genetic diversity in Clerodendrum species of North East India. Mol Biol Rep 47:7365–7377. https://doi.org/10.1007/s11033-020-05792-x
Arlos HQJ, Zulita APL (2020) Genetic variability in Pouteria lucuma using ISSR markers. Manglar 17(1):7–12. https://doi.org/10.17268/manglar.2020.002
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus. 12:13–15
Sambrook, Russell DW (2001) Molecular cloning: a Laboratory manual. Cold spring Harbour Laboratoey Press, Cold spring harbor, NY, USA
Aghaei M, Darvishzadeh R, Hassani A (2012) Molecular characterization and similarity relationships among Iranian basil (O basilicum) accessions using inter simple sequence repeat markers. Artig Cient 43(2):312–320
Sambrook J, Fritschi EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York
Mohamad A, Alhasnawi AN, Kadhimi AA, Isahak A et al (2017) DNA Isolation and Optimization of ISSR-PCR Reaction System in Oryza sativa L. Int J. Adv Sci Eng Inf Tech 7(6) ISSN: 2088-5334
Hammer Ø, Harper DAT, Ryan PD (2001) PAST: paleontological statistics software package for education and data analysis. Palaeontologia Electronica 4(1):9
Jaccard P (1908) Nouvelles recherches sur la distribution florale. Bull. Soc. Vaud. Sci. Nat. 44:223–270
Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci 76(10):5269–5273. https://doi.org/10.1073/pnas.76.10.5269
Ruiz R, Gilliland TJ, Calsyn E, Gillilad TJ, Coll R, Van Eijk MJT, De Loose M (2000) Estimating genetic conformity between related ryegrass (Lolium) varieties II. AFLP characterization. Mol Breed 6:593–602
Soengas P, Velasco P, Padilla G, Ordas A, Cartea ME (2006) Genetic relationships among brassica napus crops based on SSR marker. Am Soc Horticul Sci 41(5):1195–1199. https://doi.org/10.21273/HORTSCI.41.5.1195
Mondini L, Noorani A, Pagnotta MA (2009) Assessing plant genetic diversity by molecular tools. Diversity. 1:19–35. https://doi.org/10.3390/d1010019
Nagaraju J, Reddy K, Nagaraja G et al (2001) Comparison of multilocus RFLPs and PCR-based marker systems for genetic analysis of the silkworm, Bombyx mori. Heredity 86:588–597. https://doi.org/10.1046/j.1365-2540.2001.00861.x
Zhiqiang M, Zhang C, Khan MA et al (2015) Efficiency of improved RAPD and ISSR markers in assessing genetic diversity and relationships in Angelica sinensis Oliv. Diels varieties of China. Electron J Biotechnol 18(2):96–102. https://doi.org/10.1016/j.ejbt.2014.12.006
Mandal AB, Thomas VA, Elanchezhian R (2007) RAPD pattern of Costus speciosus Koen ex. Retz., an important medicinal plant from the Andaman and Nicobar Islands. Curr Sci 933:369–373
Samantaray S, Ngangkham U (2017) Evaluation of genetic diversity in Chlorophytum borivilianum (Santp. and Fernan.) using molecular markers; an endangered medicinal plant. Active ingredients from Aromat Med Plants 50-70
Jayaram K, Prasad MNV (2008) Genetic diversity in Oroxylum indicum L. Vent. Bignoniaceae, a vulnerable medicinal plant by random amplified polymorphic DNA marker. Afr J Biotech 7:254–262
Pei YL, Zou YP, Yin Z, Wang XQ, Zhang ZX, Hong DY (1995) Preliminary report of RAPD analysis in Paeonia suffruticosa subsp. Spontanea and P. rockii. Acta Phys Sin 33:350–356
Brauner S, Crawford DJ, Stuessy TF (1992) Ribosomal DNA and RAPD variation in the rare plant family Lactoridaceae. Am J Bot 79:1436–1439
Xiaoyan Liu DuJ, Khan MA, Cheng J, Wei C et al (2020) Analysis of genetic diversity and similarities between different Lycium varieties based on ISSR analysis and RAMP-PCR markers. World Acad Sci J. https://doi.org/10.3892/wasj.2020.39
Jingang W, Ying G, Daidi C, Shenkui L, Chuanpin Y (2008) ISSR analysis of 26 general species of Gladiolus hybridus Hort. J Northeast Agric Univ 8(154):6–10
Chaudhary V, Kumar M, Sharma S et al (2018) (2018) Assessment of genetic diversity and population structure in gladiolus (Gladiolus hybridus Hort) by ISSR markers. Physiol Mol Biol Plants 24:493–501. https://doi.org/10.1007/s12298-018-0519-2
Verma KS, Haq SU, Kachhwaha S, Kothari SL (2017) RAPD and ISSR marker assessment of genetic diversity in Citrullus colocynthis L. Schrad: a unique source of germplasm highly adapted to drought and high temperature stress. 3 Biotech 75:288. https://doi.org/10.1007/s13205-017-0918-z
Nader RA, Hayssam MA (2020) Mohamed ZMS and Hosam EEW (Quantitative and Qualitative Genetic Studies of Some Acacia Species Grown in Egypt. Plants 9(2):243. https://doi.org/10.3390/plants9020243
Dasgupta N, Nandy P, Sengupta C, Das S (2015) RAPD and ISSR marker mediated genetic polymorphism of two mangroves Bruguiera gymnorrhiza and Heritiera fomes from Indian Sundarbans in relation to their sustainability. Physiol Mol Biol Plants 21(3):375–384. https://doi.org/10.1007/s12298-015-0308-0
Farajpour M, Ebrahimi M, Amiri R et al (2011) Study of genetic variation in yarrow using inter-simple sequence repeat (ISSR) and random amplified polymorphic DNA (RAPD) markers. Afr J Biotechnol 10(54):11137–11141
Patel DM, Fougat RS, Sakure AA, Kumar S, Kumar M, Mistry JG (2016) Detection of genetic variation in sandalwood using various DNA markers. 3 Biotech 61:55.
Tripathi N, Chouhan DS, Saini N, Tiwari S (2012) Assessment of genetic variations among highly endangered medicinal plant Bacopa monnieri L. from Central India using RAPD and ISSR analysis. 3 Biotech 24:327–336. https://doi.org/10.1007/s13205-012-0059-3
Shafie MS, Hasan SM, Zain AM, Shah RM (2011) RAPD and ISSR markers for comparative analysis of genetic diversity in wormwood capillary Artemisia capillaris from Negeri Sembilan, Malaysia. J Med Plants Res 518:4426–4451
Preethi P, Rahman S, Naganeeswaran S et al (2020) Development of EST-SSR markers for genetic diversity analysis in coconut (Cocos nucifera L.). Mol Biol Rep. https://doi.org/10.1007/s11033-020-05981-8
Taran B, Zhang C, Warkentin T, Tullu A, Vanderberg A (2005) Genetic diversity among varieties and wild species accessions of pea Pisum sativum L. based on molecular markers, and morphological and physiological characters. Genome 48:257–272. https://doi.org/10.1139/g04-114
Velasco-Ramirez AP, Torres-Moran MI, Molina-Moret S et al (2014) Efficiency of RAPD, ISSR, AFLP and ISTR markers for the detection of polymorphisms and genetic relationships in camote de cerro Dioscorea spp. Electron J Biotechnol 172:65–71. https://doi.org/10.1016/j.ejbt.2014.01.002
Debajit S, Sukriti D, Sneha G, Mohan L et al (2015) RAPD and ISSR based Intra-specific molecular genetic diversity analysis of Cymbopogon flexuosus L. Stapf with a distinct correlation of morpho-chemical observations. Res J Biotechnol 107:105–113
Acknowledgements
The authors are sincerely thankful to the Director General, Madhya Pradesh Council of Science and Technology, Vigyan Bhawan, Nehru Nagar, for providing all the necessary facilities to conduct the research. We also thank Dr SS Asthana for editing the Manuscript. We also thank our reviewers for improving the earlier draft by their constructive comments to finalize the manuscript.
Author information
Authors and Affiliations
Contributions
Laboratory experiment work was performed by AT with the help of laboratory staff. AT has analyzed the data, written and critically reviewed the manuscript. Collection of samples was done by AT with the help of AK from Different parts of India. Facilities in the lab are provided by RS.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they do not have any conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Tilwari, A., Sharma, R. Random amplified polymorphic DNA and inter simple sequence repeat markers reveals genetic diversity between micro propagated, wild and field cultivated genotypes of Gloriosa superba: an endangered medicinal plant. Mol Biol Rep 48, 2437–2452 (2021). https://doi.org/10.1007/s11033-021-06278-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11033-021-06278-0