Abstract
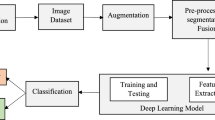
The tremendous research towards medical health systems are giving ample scope for the computing systems to emerge with the latest innovations. These innovations are leading to the efficient implementations of the medical systems which involve in automatic diagnosis of the health related problems. The most important health research is going on towards cancer prediction, which has different forms and can be affected on different portions of the body parts. One of the most affected cancer that predicted to be incurable are Pancreatic Cancer, which cannot be treated efficiently once identified, in most of the cases it found to be unpredictable as it lies in the abdomen region below the stomach. Therefore the advancements in the medical research is trending towards the implementations of an automated systems which identifies the stages of cancer if affected and provide the better diagnosis and treatment if identified. Deep learning is one such area which extended its research towards medical imaging, which automates the process of diagnosing the problems of the patients when appended with the set of machines like CT/PET Scan systems. In this paper, the deep learning strategy named Convolutional Neural network (CNN) model is used to predict the cancer images of the pancreas, which is embedded with the model of Gaussian Mixture model with EM algorithm to predict the essential features from the CT Scan and predicts the percentage of cancer spread in the pancreas with the threshold parameters taken as a markers. The experimentation is carried out on the CT Scan images dataset of pancreas collected from the Cancer Imaging Archive (TCIA) consists of approximately 19,000 images supported by the National Institutes of Health Clinical Center to analyze the performance of the model.









Similar content being viewed by others
References
Arevaloa J, Gonzáleza FA, Ramos-Pollánb R, Oliveirac JL, Guevara Lopez MA (2016) Representation learning for mammography mass lesion classification with convolutional neural networks. Comput Methods Prog Biomed 127:248–257
Asri H, Mousannif H, Al Moatassime H, Noël T (2016) Using Machine Learning Algorithms for Breast Cancer Risk Prediction and Diagnosis. Procedia Computer Science 83:1064–1069
Bridge CP et al (2018) Fully-automated analysis of body composition from CT in cancer patients using convolutional neural networks. In: Stoyanov D. et al. (eds) OR 2.0 Context-Aware Operating Theaters, Computer Assisted Robotic Endoscopy, Clinical Image-Based Procedures, and Skin Image Analysis. CARE 2018, CLIP 2018, OR 2.0 2018, ISIC 2018. Lecture Notes in Computer Science, vol 11041. Springer
Ciresan DC, Giusti A, Gambardella LM, Schmidhuber J (2013) Mitosis detection in breast cancer histology images with deep neural networks. In: International Conference on Medical Image Computing and Computer Assisted Intervention. Springer, pp. 411–418
Germán Corredor, Germán Corredor, Xiangxue Wang, Xiangxue Wang, Cheng Lu, Cheng Lu, Vamsidhar Velcheti, Vamsidhar Velcheti, Eduardo Romero, Eduardo Romero, Anant Madabhushi, Anant Madabhushi (2018) A watershed and feature-based approach for automated detection of lymphocytes on lung cancer images. Proc. SPIE 10581, Medical Imaging 2018: Digital Pathology, 105810R
Gerasa KJ, Wolfsonc S, Kimc SG, Moyc L, Cho K (2017) High-resolution breast cancer screening with multi-view deep convolutional neural networks. arXiv:1703.07047v1
Glicksberg BS, Miotto R, Johnson KW, Shameer K, Li L, Chen R, Dudley JT (2018) Automated disease cohort selection using word embeddings from Electronic Health Records. Pac Symp Biocomput
Hussein S, Kandel P, Bolan CW, Wallace MB, Bagci U (2019) Lung and pancreatic tumor characterization in the deep learning era: novel supervised and unsupervised learning approaches. IEEE Transactions on Medical Imaging, ISSN:0278–0062
Isola P, Zhu J, Zhou T, Efros A (2016) Image-to-Image Translation with Conditional Adversarial Networks. arXiv:1611.07004v1
Li H, Zhong H, Boimel PJ, Ben-Josef E, Xiao Y, Fan Y (2017) Deep convolutional neural networks for imaging based survival analysis of rectal cancer patients. International Journal of Radiation Oncology 99
Miotto R, Li L, Dudley JT (2016) Deep learning to predict patient future diseases from the electronic health records. European Conference on Information Retrieval
Paeng K, Hwang S, Park S, Kim M, Kim S (2016) A unified framework for tumor proliferation score prediction in breast histopathology. arXiv:1612.07180v1
Pearce C (2018) Convolutional neural networks and the analysis of cancer imagery. Stanford University
Radford A, Jozefowicz R, Sutskever I (2017) Learning to generate reviews and discovering sentiment. arXiv:1704.01444, https://arxiv.org/pdf/1704.01444.pdf
Rubadue C, Suster D, Wang D (2016) Deep learning assessment of tumor proliferation in breast cancer histological images. arXiv:1610.03467. https://arxiv.org/pdf/1610.03467.pdf
Shelhamer E, Long J, Darrell T (2016) Fully convolutional networks for semantic segmentation. arXiv:1605.06211v1
Veta M, van Diest PJ, Jiwa M, Al-Janabi S, Pluim JP (2016) Mitosis counting in breast cancer: Object-level interobserver agreement and comparison to an automatic method. PLoS One 11(8):e0161286
Zhen X, Chen J, Zhong Z, Hrycushko B, Zhou L, Jiang S, Albuquerque K, Gu X (2017) Deep convolutional neural network with transfer learning for rectum toxicity prediction in cervical cancer radiotherapy: a feasibility study. Institute of Physics and Engineering in Medicine Physics in Medicine & Biology 62
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sekaran, K., Chandana, P., Krishna, N.M. et al. Deep learning convolutional neural network (CNN) With Gaussian mixture model for predicting pancreatic cancer. Multimed Tools Appl 79, 10233–10247 (2020). https://doi.org/10.1007/s11042-019-7419-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11042-019-7419-5