Abstract
Introduction
Glioblastoma (GBM) is heterogeneous and underlying genomic profiles influence evolution, resistance, and therapeutic responses. While extensive knowledge regarding genomic profiling of primary GBM exists, there remains a lack of understanding of genomic differences in recurrent GBM.
Methods
We used the FoundationOne® comprehensive genomic profiling assay (CGP) to analyze ten matched primary and recurrent GBM. Genomic alterations (GA) were compared to the cancer database Catalogue of Somatic Mutations in Cancer (COSMIC).
Results
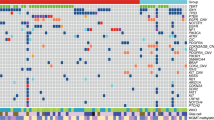
All matched tumor pairs demonstrated differences in GA between the primary and recurrence including one resected without any intervening therapy. This suggests that time and/or therapeutic intervention contribute to GA. Although mutations were common to both the primary and recurrence, the percent reads varied substantially suggesting clonal expansions and contractions. For example, EGFR mutations were significantly expanded in three patients, and CNAs were increased in two patients at recurrence. Four genes that were commonly altered in both primary and recurrent GBM were more prevalent in our cohort than reported in COSMIC: CDKN2A (86% vs. 53%) and CDKN2B (86% vs. 54%) deletions, EGFR activating mutation (52% vs. 10%) or amplification (81% vs. 45%), and TERT mutation (95% vs. 51%). Lastly, PI3K pathway activating mutations were also commonly seen in our cohort (67%).
Conclusions
CGP revealed that GA identified in GBM changed over time and with treatment. Mutations in TERT, CDKN2A/CDKN2B, EGFR, and PI3K pathway were commonly observed in both primary and recurrent GBM revealing their prognostic and therapeutic potential. This may have important implications for individualized therapies and needs further evaluation.




Similar content being viewed by others
References
Marusyk A, Almendro V, Polyak K (2012) Intra-tumour heterogeneity: a looking glass for cancer? Nat Rev Cancer 12:323–334. https://doi.org/10.1038/nrc3261
Marusyk A, Polyak K (2010) Tumor heterogeneity: causes and consequences. Biochim Biophys Acta 1805:105–117. https://doi.org/10.1016/j.bbcan.2009.11.002
Suzuki H, Aoki K, Chiba K, Sato Y, Shiozawa Y, Shiraishi Y, Shimamura T, Niida A, Motomura K, Ohka F, Yamamoto T, Tanahashi K, Ranjit M, Wakabayashi T, Yoshizato T, Kataoka K, Yoshida K, Nagata Y, Sato-Otsubo A, Tanaka H, Sanada M, Kondo Y, Nakamura H, Mizoguchi M, Abe T, Muragaki Y, Watanabe R, Ito I, Miyano S, Natsume A, Ogawa S (2015) Mutational landscape and clonal architecture in grade II and III gliomas. Nat Genet 47:458–468. https://doi.org/10.1038/ng.3273
Yap TA, Gerlinger M, Futreal PA, Pusztai L, Swanton C (2012) Intratumor heterogeneity: seeing the wood for the trees. Sci Transl Med 4:127ps110. https://doi.org/10.1126/scitranslmed.3003854
Sattiraju A, Sai KKS, Mintz A (2017) Glioblastoma stem cells and their microenvironment. Adv Exp Med Biol 1041:119–140. https://doi.org/10.1007/978-3-319-69194-7_7
Campbell LL, Polyak K (2007) Breast tumor heterogeneity: cancer stem cells or clonal evolution? Cell Cycle 6:2332–2338. https://doi.org/10.4161/cc.6.19.4914
Orzan F, De Bacco F, Crisafulli G, Pellegatta S, Mussolin B, Siravegna G, D’Ambrosio A, Comoglio PM, Finocchiaro G, Boccaccio C (2017) Genetic evolution of glioblastoma stem-like cells from primary to recurrent tumor. Stem Cells. https://doi.org/10.1002/stem.2703
O’Connor ML, Xiang D, Shigdar S, Macdonald J, Li Y, Wang T, Pu C, Wang Z, Qiao L, Duan W (2014) Cancer stem cells: a contentious hypothesis now moving forward. Cancer Lett 344:180–187. https://doi.org/10.1016/j.canlet.2013.11.012
Greaves M, Maley CC (2012) Clonal evolution in cancer. Nature 481:306–313. https://doi.org/10.1038/nature10762
Consortium G (2018) Glioma through the looking GLASS: molecular evolution of diffuse gliomas and the glioma longitudinal analysis consortium. Neuro Oncol 20:873–884. https://doi.org/10.1093/neuonc/noy020
Mazor T, Pankov A, Johnson BE, Hong C, Hamilton EG, Bell RJA, Smirnov IV, Reis GF, Phillips JJ, Barnes MJ, Idbaih A, Alentorn A, Kloezeman JJ, Lamfers MLM, Bollen AW, Taylor BS, Molinaro AM, Olshen AB, Chang SM, Song JS, Costello JF (2015) DNA methylation and somatic mutations converge on the cell cycle and define similar evolutionary histories in brain tumors. Cancer Cell 28:307–317. https://doi.org/10.1016/j.ccell.2015.07.012
Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD, Kleihues P, Ellison DW (2016) The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol 131:803–820. https://doi.org/10.1007/s00401-016-1545-1
Kim H, Zheng S, Amini SS, Virk SM, Mikkelsen T, Brat DJ, Grimsby J, Sougnez C, Muller F, Hu J, Sloan AE, Cohen ML, Van Meir EG, Scarpace L, Laird PW, Weinstein JN, Lander ES, Gabriel S, Getz G, Meyerson M, Chin L, Barnholtz-Sloan JS, Verhaak RG (2015) Whole-genome and multisector exome sequencing of primary and post-treatment glioblastoma reveals patterns of tumor evolution. Genome Res 25:316–327. https://doi.org/10.1101/gr.180612.114
Szerlip NJ, Pedraza A, Chakravarty D, Azim M, McGuire J, Fang Y, Ozawa T, Holland EC, Huse JT, Jhanwar S, Leversha MA, Mikkelsen T, Brennan CW (2012) Intratumoral heterogeneity of receptor tyrosine kinases EGFR and PDGFRA amplification in glioblastoma defines subpopulations with distinct growth factor response. Proc Natl Acad Sci USA 109:3041–3046. https://doi.org/10.1073/pnas.1114033109
Blumenthal DT, Dvir A, Lossos A, Tzuk-Shina T, Lior T, Limon D, Yust-Katz S, Lokiec A, Ram Z, Ross JS, Ali SM, Yair R, Soussan-Gutman L, Bokstein F (2016) Clinical utility and treatment outcome of comprehensive genomic profiling in high grade glioma patients. J Neuro-Oncol 130:211–219. https://doi.org/10.1007/s11060-016-2237-3
Frattini V, Trifonov V, Chan JM, Castano A, Lia M, Abate F, Keir ST, Ji AX, Zoppoli P, Niola F, Danussi C, Dolgalev I, Porrati P, Pellegatta S, Heguy A, Gupta G, Pisapia DJ, Canoll P, Bruce JN, McLendon RE, Yan H, Aldape K, Finocchiaro G, Mikkelsen T, Prive GG, Bigner DD, Lasorella A, Rabadan R, Iavarone A (2013) The integrated landscape of driver genomic alterations in glioblastoma. Nat Genet 45:1141–1149. https://doi.org/10.1038/ng.2734
Johnson BE, Mazor T, Hong C, Barnes M, Aihara K, McLean CY, Fouse SD, Yamamoto S, Ueda H, Tatsuno K, Asthana S, Jalbert LE, Nelson SJ, Bollen AW, Gustafson WC, Charron E, Weiss WA, Smirnov IV, Song JS, Olshen AB, Cha S, Zhao Y, Moore RA, Mungall AJ, Jones SJM, Hirst M, Marra MA, Saito N, Aburatani H, Mukasa A, Berger MS, Chang SM, Taylor BS, Costello JF (2014) Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science 343:189–193. https://doi.org/10.1126/science.1239947
Muscat AM, Wong NC, Drummond KJ, Algar EM, Khasraw M, Verhaak R, Field K, Rosenthal MA, Ashley DM (2018) The evolutionary pattern of mutations in glioblastoma reveals therapy-mediated selection. Oncotarget 9:7844–7858. https://doi.org/10.18632/oncotarget.23541
Schonberg DL, Bao S, Rich JN (2013) Genomics informs glioblastoma biology. Nat Genet 45:1105–1107. https://doi.org/10.1038/ng.2775
Hayashi Y, Ueki K, Waha A, Wiestler OD, Louis DN, von Deimling A (1997) Association of EGFR gene amplification and CDKN2 (p16/MTS1) gene deletion in glioblastoma multiforme. Brain Pathol 7:871–875
Moulton T, Samara G, Chung WY, Yuan L, Desai R, Sisti M, Bruce J, Tycko B (1995) MTS1/p16/CDKN2 lesions in primary glioblastoma multiforme. Am J Pathol 146:613–619
Ueki K, Ono Y, Henson JW, Efird JT, von Deimling A, Louis DN (1996) CDKN2/p16 or RB alterations occur in the majority of glioblastomas and are inversely correlated. Cancer Res 56:150–153
Schlegel J, Stumm G, Brandle K, Merdes A, Mechtersheimer G, Hynes NE, Kiessling M (1994) Amplification and differential expression of members of the erbB-gene family in human glioblastoma. J Neuro-Oncol 22:201–207
Nonoguchi N, Ohta T, Oh JE, Kim YH, Kleihues P, Ohgaki H (2013) TERT promoter mutations in primary and secondary glioblastomas. Acta Neuropathol 126:931–937. https://doi.org/10.1007/s00401-013-1163-0
Frampton GM, Fichtenholtz A, Otto GA, Wang K, Downing SR, He J, Schnall-Levin M, White J, Sanford EM, An P, Sun J, Juhn F, Brennan K, Iwanik K, Maillet A, Buell J, White E, Zhao M, Balasubramanian S, Terzic S, Richards T, Banning V, Garcia L, Mahoney K, Zwirko Z, Donahue A, Beltran H, Mosquera JM, Rubin MA, Dogan S, Hedvat CV, Berger MF, Pusztai L, Lechner M, Boshoff C, Jarosz M, Vietz C, Parker A, Miller VA, Ross JS, Curran J, Cronin MT, Stephens PJ, Lipson D, Yelensky R (2013) Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol 31:1023–1031. https://doi.org/10.1038/nbt.2696
Forbes SA, Beare D, Boutselakis H, Bamford S, Bindal N, Tate J, Cole CG, Ward S, Dawson E, Ponting L, Stefancsik R, Harsha B, Kok CY, Jia M, Jubb H, Sondka Z, Thompson S, De T, Campbell PJ (2017) COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res 45:D777–D783. https://doi.org/10.1093/nar/gkw1121
Ohgaki H, Kleihues P (2013) The definition of primary and secondary glioblastoma. Clin Cancer Res 19:764–772. https://doi.org/10.1158/1078-0432.CCR-12-3002
Felsberg J, Hentschel B, Kaulich K, Gramatzki D, Zacher A, Malzkorn B, Kamp M, Sabel M, Simon M, Westphal M, Schackert G, Tonn JC, Pietsch T, von Deimling A, Loeffler M, Reifenberger G, Weller M, German Glioma N (2017) Epidermal growth factor receptor variant III (EGFRvIII) positivity in EGFR-amplified glioblastomas: prognostic role and comparison between primary and recurrent tumors. Clin Cancer Res 23:6846–6855. https://doi.org/10.1158/1078-0432.CCR-17-0890
Kim J, Lee IH, Cho HJ, Park CK, Jung YS, Kim Y, Nam SH, Kim BS, Johnson MD, Kong DS, Seol HJ, Lee JI, Joo KM, Yoon Y, Park WY, Lee J, Park PJ, Nam DH (2015) Spatiotemporal Evolution of the Primary Glioblastoma Genome. Cancer Cell 28:318–328. https://doi.org/10.1016/j.ccell.2015.07.013
Meric-Bernstam F, Frampton GM, Ferrer-Lozano J, Yelensky R, Perez-Fidalgo JA, Wang Y, Palmer GA, Ross JS, Miller VA, Su X, Eroles P, Barrera JA, Burgues O, Lluch AM, Zheng X, Sahin A, Stephens PJ, Mills GB, Cronin MT, Gonzalez-Angulo AM (2014) Concordance of genomic alterations between primary and recurrent breast cancer. Molecular Cancer Ther 13:1382–1389. https://doi.org/10.1158/1535-7163.MCT-13-0482
Vitucci M, Hayes DN, Miller CR (2011) Gene expression profiling of gliomas: merging genomic and histopathological classification for personalised therapy. Br J Cancer 104:545–553. https://doi.org/10.1038/sj.bjc.6606031
Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, Misra A, Nigro JM, Colman H, Soroceanu L, Williams PM, Modrusan Z, Feuerstein BG, Aldape K (2006) Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 9:157–173. https://doi.org/10.1016/j.ccr.2006.02.019
Acknowledgements
We acknowledge Foundation Medicine for performing the FoundationOne® comprehensive genomic profiling in this investigator initiated study.
Funding
This work was supported by NCI F30 CA203397 to B.K.N. The funders had no role in the study design, data collection and interpretation, or the decision to submit the work for publication.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors. This study was deemed exempt by the UNMC Institutional Review Board and the Fred and Pamela Buffet Cancer Center Scientific Review Committee as all patients included are deceased. Approval for this study, including a waiver of informed consent and a HIPAA waiver of authorization, was also obtained from the Western Institutional Review Board (Protocol No. 20152817).
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Neilsen, B.K., Sleightholm, R., McComb, R. et al. Comprehensive genetic alteration profiling in primary and recurrent glioblastoma. J Neurooncol 142, 111–118 (2019). https://doi.org/10.1007/s11060-018-03070-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11060-018-03070-2