Abstract
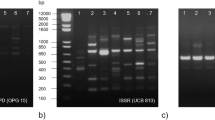
Moringa oleifera Lam. (drumstick) belongs to the family Moringaceae that is originated from sub-Himalayan tracts of Northern India distributed worldwide in the tropics and sub-tropics. Immature pods and fresh leaves are widely used as vegetable and are rich source of minerals and vitamins. In the present work, we made an attempt to develop and use a set of RAPD-SCAR marker for the identification of superior germplasm of M. oleifera from the accessions collected from South India. Initially, 120 trees were surveyed based on total fruit yield, and single fruit weight from Karnataka, Kerala, and Tamil Nadu states of India; 23 plants had 50% higher fruit yield and single fruit weight than average and were selected as Candidate Plus Trees (CPTs). On the basis of morphological and biochemical analysis, CPT17 was selected as elite germplasm. Random amplified polymorphic DNA (RAPD) analysis of CPTs indicated 89.61% polymorphism among 23 CPTs. These markers could be used in marker-assisted selection and breeding programs in M. oleifera. Further, an attempt to develop a set of RAPD-SCAR marker for the identification of superior germplasm of M. oleifera was made. RAPD primer OPA-19 (CAAACGTCGG) revealed a unique band (1500-bp) in CPT17. The specific RAPD band was recovered from the gel, cloned, and sequenced. BLAST analysis of the CPT17 specific sequences revealed that no considerable similarity with known protein. Based on these unique characterized sequences, specific primers for CPT17 were designed. Specific amplification profile of this primer proved it as a SCAR marker (F2R2) for CPT17 genotype.



Similar content being viewed by others
References
Abraham G, Pandey N, Mishra V, Chaudhary AA, Ahmad A, Singh R, Singh PK (2013) Development of SCAR based molecular markers for identification of different species of Azolla. Indian J Biotechnol 12:489–492
Amicucci A, Rossi I, Potenza L, Agostini D, Stocchi V (1997) Use of sequence characterised amplified region and RAPD markers in the identification of the white truffle Tuber magnatum Pico. Biotechnol Tech 11(3):149–154. https://doi.org/10.1023/A:1018493111804
Anuntalabhochai S, Sitthiphrom S, Thongtaksin W, Sanguansermsri M, Cutler RW (2007) Hybrid detection and characterization of Curcuma species using sequence characterized DNA markers. Sci Hortic 111:389–393. https://doi.org/10.1016/j.scienta.2006.11.008
Anwar F, Latif S, Ashraf M, Gilani AH (2007) Moringa oleifera: A food plant with multiple medicinal uses. Phytother Res 21(1):17–25. https://doi.org/10.1002/ptr.2023
Basha SD, Sujatha M (2007) Inter and intra-population variability of Jatropha curcas (L.) characterized by RAPD and ISSR markers and development of population-specific SCAR markers. Euphytica 156:271–431. https://doi.org/10.1007/s10681-007-9387-5
Bhagyawant SS (2016) RAPD-SCAR markers: an interface tool for authentication of traits. J Biosci Med 4:1–9. https://doi.org/10.4236/jbm.2016.41001
Busconi M, Sebastiani L, Fogher C (2006) Development of SCAR markers for germplasm characterisation in olive tree (Olea europea L.). Molecular Breeding 17: 59–68. https://doi.org/10.1007/s11032-005-1395-3
Cheng J, Long Y, Khan MA, Wei C, Fu S, Fu J (2015) Development and significance of RAPD-SCAR markers for the identification of Litchi chinensis Sonn. by improved RAPD amplification and molecular cloning. Electron J Biotechnol 18:35–39. https://doi.org/10.1016/j.ejbt.2014.11.004
Cunha CMS, Hinz RH, Pereira A, Tcacenco FA (2015) A SCAR marker for identifying susceptibility to Fusarium oxysporum f. sp. cunense in banana. Scientia Horticulturae 191: 108–112. https://doi.org/10.1016/j.scienta.2015.04.038
Dahlberg JA, Zhang X, Hart GE, Mullet JE (2002) Comparative assessment of variation among sorghum germplasm accessions using seed morphology and RAPD measurements. Crop Science 42: 291–296. https://doi.org/10.2135/cropsci2002.0291
Dnyaneshwar W, Preeti C, Kalpana J, Bhushan P (2006) Development and application of RAPD-SCAR marker for identification of Phyllanthus emblica L. Biol Pharm Bull 29:2313–2316. https://doi.org/10.1248/bpb.29.2313
Duke JA (1983) Moringa oleifera Lam. Handbook of energy crops. Center for New Crops & Plants Products. Purdue University.[Consultado 28 de mayo de2013] http://www.hort.purdue.edu/newcrop/duke_energy/Moringa_oleifera.html
Durai M, Dubey SC, Triphathi A (2012) Genetic diversity analysis and development of scar marker for detection of Indian populations of Fusarium oxysporum f. sp. ciceris causing chickpea wilt. Folia Microbiology 57:229–235. https://doi.org/10.1007/s12223-012-0118-5
Edwards S, Tadesse M, Demissew S, Hedberg I (2000) Flora of Ethiopia and Eritrea, Vol. II part 1: Magnoliaceae to Flacourtiaceae. Addis Ababa, Ethiopia and Uppsala, Sweden: The National Herbarium, Addis Ababa University p532
Ellsworth DL, Rittenhouse KD, Honeycutt RL (1993) Artifactual variation in Random amplified polymorphic DNA banding patterns. Biotechniques 14:214–217
Ferreira ME, Grattapaglia D (1996) Introducaoaouso de marcadoresmolecularesemanalisegenetica. Brasilia:Embrapa-Cenargen p 220
Innis MA, Gelfand DH, Sninsky JJ, White TJ (1990) PCR protocols: a guide to methods and application. San Diego:Academic Press p 482
Kim H, Han D, Kang J, Choi1 Y, Levi A, Pyo GL, Park Y (2015) Sequence-characterized amplified polymorphism markers for selecting rind stripe pattern in watermelon (Citrullus lanatus L.). Horticulture, Environment and Biotechnology 56(3): 341–349. https://doi.org/10.1007/s13580-015-0017-1
Kiran U, Khan S, Mirza KJ, Ram M, Abdin MZ (2010) SCAR markers: a potential tool for authentication of herbal drugs. Fitoterapia 81:969–976. https://doi.org/10.1016/j.fitote.2010.08.002
Korekar G, Sharma RK, Kumar R, Bisht NC, Srivastava RB, Ahuja PS, Stobdan T (2012) Identification and validation of sex-linked SCAR markers in dioecious Hippophae rhamnoides L. (Elaeagnaceae). Biotech Letter 34:973–978. https://doi.org/10.1007/s10529-012-0852-4
Lee MY, Doh EJ, Chae HP, Young HK, Eung SK, Ko BS (2006) Developmentof SCAR marker for discrimination of A. princeps and A. argyi from other Artemisia Herbs. Biology and Pharmaceutical Bulletin 29:629–630. https://doi.org/10.1248/bpb.29.629
Li G, Park YJ (2012) SCAR Markers for discriminating species of two genera of medicinal plants, Liriope and Ophiopogon. Genet Mol Res 11:2987–2995. https://doi.org/10.4238/2012.May.18.14
Li H, Zhang SG, Gao JM, Wang CG, Zhang Y, Qi LW, Chen L, Song WQ (2008) Development of a sequence characterized amplified region (SCAR) marker associated with high rooting ability in Larix. Biol Plant 52(3):525–528. https://doi.org/10.1007/s10535-008-0102-8
Li HB, Wu XQ, Peng HZ, Fu LZ, Wei HL, Wu QQ, Jin QY, Li N (2008) New available SCAR markers: potentially useful in distinguishing a commercial strain of the superior type from other’s trains of Lentinula edodes in China. App Microb Biotech 81: 303–309. https://doi.org/10.1007/s00253-008-1671-3
Litt M, Luty JA (1989) A hypervariable microsatellite revealed by in vitro amplification of a dinucleotide repeat within the cardiac muscle actin gene. Am J Hum Genet 44:397–401
Marieschi M, Torelli A, Beghe D, Bruni R (2016) Authentication of Punica granatum L.: Development of SCAR markers for the detection of 10 fruits potentially used in economically motivated adulteration. Food Chem 202:438–444. https://doi.org/10.1016/j.foodchem.2016.02.011
Mezei LM, Storts DR (1994) Purification of PCR products. In: Griffin HG, Griffin AM (eds) PCR Technology: Current Innovations. CRC Press, Boca Raton, FL, p 21
Mughal MH, Ali G, Srivastava PS, Iqbal M (1999) Improvement of drumstick (Moringa pterygosperma Gaertn.)—a unique source of food and medicine through tissue culture. Hamdard Medicine 42:37–42
Muralidharan K, Wakeland EK (1993) Concentration of primer and template qualitatively affects products in RAPD-PCR. Biotechniques 14:362–364
Navie S, Csurhes S (2011) Weed risk assessment horseradish tree Moringa oleifera. Biosecurity Queensland Department of Employment, Economic Development and Innovation, Brisbane 4001:1–22
Nkongolo KK, Michael P, Gratton WS (2002) Identification and characterization of RAPD markers inferring genetic relationships among Pine species. Genome 45(1):51–58. https://doi.org/10.1139/g01-121
Olson ME (2002) Combining data from DNA sequences and morphology for a phylogeny of Moringaceae (Brassicales). Syst Bot 27:55–73. https://doi.org/10.1043/0363-6445-27.1.55
Paran I, Michelmore RW (1993) Development of reliable PCR-based markers linked to downy mildew resistance genes in lettuce. Theor Appl Genet 85:985–993. https://doi.org/10.1007/BF00215038
Palada MC, Ebert AW (2012) Moringa. In: PeterKV HP (ed) Handbook of Vegetables. Stadium Press, New Delhi, India, pp 193–242
Parasnis AS, Gupta VS, Tamhankar SA, Ranjekar PK (2000) A highly reliable sex diagnostic PCR assay for mass-screening of Papaya seedlings. Mol Breeding 6:337–344. https://doi.org/10.1023/A:1009678807507
Qaiser M (1973) Moringaceae. In flora of West Pakistan. Nasir E and Ali SI pp 1–4
Ravi RSD, Siril EA, Nair BR (2020) Proximate and nutritive value screening of Moringa oleifera, a multipurpose tree, and identification of elite germplasm. Abrahamia 6:16–25
Robles J, Doers M (1994) pGEM®-T Vector Systems troubleshooting guide. Promega Notes 45:19–20
Saiki RK, Gelfand DH, Stoffel S, Scharf SJ, Higuchi R, Horn GT, Mullis KB, Erlich HA (1988) Primer directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 239: 487–491. https://doi.org/10.1126/science.2448875
Saini RK, Saad KR, Ravishankar GA, Giridhar P, Shetty NP (2013) Genetic diversity of commercially grown Moringa oleifera Lam. cultivars from India by RAPD, ISSR and cytochrome P450-based markers. Plant Syst Evol 299:1205–1213. https://doi.org/10.1007/s00606-013-0789-7
Saini RK, Shetty NP, Giridhar P, Ravishankar GA (2012) Rapid in vitro regeneration method for Moringa oleifera and performance evaluation of field grown utritionally enriched tissue cultured plants. 3 Biotech 2: 187–192. https://doi.org/10.1007/s13205-012-0045-9
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, USApp 6–11
Semsang N, Chundet R, Phanchisri B (2013) Development of a SCAR marker for discrimination of a Thaijasmine rice (Oryza sativa L. cv. KDML105) mutant, BKOS6, and associated with purple color trait in Thai jasmine rice-related varieties. American Journal of Plant Sciences 4: 1774–1783. https://doi.org/10.4236/ajps.2013.49218
Smith JSC, Register JC (1998) Genetic purity and testing technologies for seed quality: a company perspective. Seed Sci Res 8(5):285–293. https://doi.org/10.1017/S0960258500004189
Solieri L, Giudici P (2010) Development of a sequence-characterized amplified region marker-targeted quantitative PCR assay for strain-specific detection of Oenococcus oeni during wine malolactic fermentation. Appl Environ Microbiol 76(23):7765–7774. https://doi.org/10.1128/AEM.00929-10
Somali MA, Bajnedi MA, Al-Faimani SS (1984) Chemical composition and characteristics of Moringa peregrina seeds and seed oil. J Am Oil Chem Soc 61:85–86
Srivastava RK, Mishra SK, Singh AK, Mohapatra T (2012) Development of a coupling-phase SCAR marker linked to the powdery mildew resistance gene ‘er1’in pea (Pisum sativum L.). Euphytica 186:855–866. https://doi.org/10.1007/s10681-012-0650-z
Sujatha M, Makkar HPS, Becker K (2005) Shoot bud proliferation from axillary nodes and leaf sections of non-toxic Jatropha curcas L. Plant Growth Regul 47:83–90. https://doi.org/10.1007/s10725-005-0859-0
Tatikonda L, Wani SP, Kannan S, Beerelli N, Sreedevi TK, Hoisington DA, Devi P, Varshney RK (2009) AFLP-based molecular characterization of an elite germplasm collection of Jatropha curcas L., a biofuel plant. Plant Science 176(4): 505–513. https://doi.org/10.1016/j.plantsci.2009.01.006
Tautz D (1989) Hyper variability of simple sequences as a general source of polymorphic DNA markers. Nucleic Acid Res 17:6463–6471. https://doi.org/10.1093/nar/17.16.6463
Vanichanon A, Blake NK, Martin JM, Talbert LE (2000) Properties of sequence tagged-site primer sets influencing repeatability. Genome 43:47–52. https://doi.org/10.1139/g99-087
Vos P, Hogers R, Bleeker M, Reijans M, Lee TV, Hornes M, Friters A, Pot J, Paleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23(21):4407–4414. https://doi.org/10.1093/nar/23.21.4407
Wang PM, Wu XC, Chi XQ, Li YD, Zheng DQ, Ding R, Min H (2011) Development and application of RAPD-SCAR markers to identify intra-species hybrids of industrial Saccharomyces cerevisiae. World J Microbiol Biotechnol 27: 185–188. https://doi.org/10.1007/s11274-010-0430-7
Weber JL, May PE (1989) Abundant class of human DNA polymorphisms, which can be typed using the polymerase chain reaction. Am J Hum Genet 44:388–396
Williams JG, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV (1990) DNA polymorphism amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Research 18: 6531–6535. https://doi.org/10.1093/nar/18.22.6531
Yang L, Fu S, Khan MA, Zeng W, Fu J (2013) Molecular cloning and development of RAPD-SCAR markers for Dimocarpus longan variety authentication. Springerplus 2:1–8. https://doi.org/10.1186/2193-1801-2-501
Yuskianti V, Shiraishi S (2010) Sequence characterized amplified region (SCAR) markers in Sengon (Paraseriathes falcataria (L.) Nielsen. Hayati Journal of Biosciences 17(4): 167–172. https://doi.org/10.4308/hjb.17.4.167
Zietkiewicz E, Rafalski A, Labuda D (1994) Geome fingerprinting by simple sequence repeats (SSR) anchored polymerase chain reaction amplification. Genomics 20:176–183. https://doi.org/10.1006/geno.1994.1151
Acknowledgements
The authors are grateful to Dr. Suhara Beevy S, Professor and Head, Department of Botany, University of Kerala, for the facilities provided.
Funding
The authors received research fellowship from the University of Kerala (No. Ac.EI/A2/10625/2016-I) to undertake the present work.
Author information
Authors and Affiliations
Contributions
DRS conducted the experiments. DRS and EAS analyzed the data. DRS drafted the manuscript. EAS and BRN designed the experiments. EAS, BRN, and DRS revised the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare no competing interests.
Additional information
Highlights
• M. oleifera used for a wide variety of purpose thus regarded as a “multipurpose tree.”
• Specific marker for the identification of superior germplasm of M. oleifera.
• This is the first report regarding SCAR marker development for M. oleifera.
Rights and permissions
About this article
Cite this article
Ravi, D., Siril, E.A. & Nair, B.R. SCAR Marker Development for the Identification of Elite Germplasm of Moringa Oleifera Lam.-A Never Die Plant. Plant Mol Biol Rep 39, 850–861 (2021). https://doi.org/10.1007/s11105-021-01300-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-021-01300-y