Abstract
Knowing before harvesting how many plants have emerged and how they are growing is key in optimizing labour and efficient use of resources. Unmanned aerial vehicles (UAV) are a useful tool for fast and cost efficient data acquisition. However, imagery need to be converted into operational spatial products that can be further used by crop producers to have insight in the spatial distribution of the number of plants in the field. In this research, an automated method for counting plants from very high-resolution UAV imagery is addressed. The proposed method uses machine vision—Excess Green Index and Otsu’s method—and transfer learning using convolutional neural networks to identify and count plants. The integrated methods have been implemented to count 10 weeks old spinach plants in an experimental field with a surface area of 3.2 ha. Validation data of plant counts were available for 1/8 of the surface area. The results showed that the proposed methodology can count plants with an accuracy of 95% for a spatial resolution of 8 mm/pixel in an area up to 172 m2. Moreover, when the spatial resolution decreases with 50%, the maximum additional counting error achieved is 0.7%. Finally, a total amount of 170 000 plants in an area of 3.5 ha with an error of 42.5% was computed. The study shows that it is feasible to count individual plants using UAV-based off-the-shelf products and that via machine vision/learning algorithms it is possible to translate image data in non-expert practical information.
Similar content being viewed by others
Introduction
Crop monitoring is a critical function within the agricultural production chain. It is important for estimating yield, weed control, phenotyping, or disease detection that have a real impact on the economies of countries and the environment (Hayes and Decker 1996). Improper crop monitoring can lead to a waste of valuable resources such as water and fertilizer.
Traditionally crop monitoring required manual labour, as the field manager or the land owner still had to monitor the crops physically. This is time consuming and can be prone to human error (Li et al. 2019). During the last decade, precision agriculture (PA) has aided to decrease the manual labour on the fields by providing crop monitoring solutions that are less subjective, cost-efficient and robust.
With recent advancements in technology it has become more feasible to develop automated and non-destructive remote sensing-based approaches. The increasing availability of unmanned aerial vehicles (UAV) is a potential solution for remotely and flexible data acquisition on an agricultural fields without the manual labour that would be required traditionally (Rokhmana 2015; Sarron et al. 2018). Crop producers do not have to survey plots manually, but can deploy a UAV in order to take aerial photographs of their crops that can be further analysed to obtain information at the plant level.
The benefits of UAVs are their commercial availability and that today most of them can be flown with an auto-pilot. Moreover, UAVs can fly at lower altitudes with increased safety and at lower cost than manned aircraft achieving higher spatial resolutions. Images acquired with a UAV can also achieve a better spatial resolution than images provided by satellite services and can cover under the right weather conditions more than a few hundred ha per day (Rokhmana 2015). UAVs are already used in precision agriculture to improve profitability and productivity by providing synoptic data and task maps to farmers (Tokekar et al. 2016).
While using UAVs can be a cheaper and faster way to collect aerial data, however, without a translation of the acquired images into relevant information this data collection adds little value. The farmer has little benefit from aerial photographs without any translation to operational knowledge. By applying machine vision methods valuable information can be extracted from high-resolution photographs.
Crop and vegetation detection using UAVs has been widely used. In Hamuda et al. (2016) an overview of common crop segmentation approaches is reviewed. In Torres-Sánchez et al. (2015), the authors used UAV acquired aerial imagery to segment different herbaceous on the field. In this research, they used various segmentation and thresholding algorithms, including Otsu’s method (Otsu 1979), excess green (ExG) and the NDVI within an Object Based Image Analysis (OBIA) framework. This work though focused only in vegetation detection via images with Red, Green and Blue (RGB) channels and multispectral imagery. Senthilnath, et al. (2016) used a UAV to acquire RGB imagery of a tomato field and used spectral-spatial classification to classify the images in tomato and non-tomato. The authors argue that this approach can be extended for counting tomatoes, although additional studies were not communicated.
Being able to remotely distinguish different crops and the soil is a valuable application for agricultural practices (Montalvo et al. 2012). Nevertheless, it is fundamental in crop production to translate the vegetation detected in relevant information for crop producers, such as yield estimation, number of individual plants in a cultivar, or the number of flowers bulbs per plant. Yield estimation can be automatically executed by counting the amount of plants or by tracking the growth of plants (Hunt et al. 2010). Several studies have been reported where a UAV mounted with RGB and multispectral cameras have produced models that estimate crop yield within a satisfactory error (Cotton, Yeom et al. 2018; sugarcane, Som-ard et al. 2018). Detection of plant traits—identify the single plant canopy or a flower bulb—at the plant level using UAV imagery was found less often in literature. In Reza et al. 2019, the author applied k-means clustering to RGB images to detect the grain areas of rice plants. The yield was estimated with a relative error from 21 to 31%. Sorghum panicles have been detected using a two-step machine-learning-based image processing method where the proposed method achieved R2 of 0.84 in 52 labelled (a unit of the dataset) images and 0.56 for the complete dataset (Guo et al. 2018).
In Li et al. (2019), the authors estimated the number of seeds that germinated and grew via an unsupervised method that analysed UAV imagery and employed a random forest based approach that provides the actual delineation of the contours of single plants. They computed emergence rate, crop canopy cover, and emergence uniformity, which were compared to manual crop assessment. They investigated how the emergence rate and uniformity could be affected in various cultivars and using different nutrients inputs (Li et al. 2019). A drawback of this work is that the region of interest (ROI) was selected manually in many plot clusters instead of detecting single plants automatically in a large very-high resolution (VHR) RGB image. Moreover, the bounding box that defines the plant area was filtered with a predefined value (higher than 35 pixels). Finally, in this paper it seems that the authors focused more on the quantity and type of fertilizer used rather than on the counting algorithm. The results provided by the authors for what plant counting concerned are very limited. For instance, 80% of the dataset (total 540 images) used for training are labelled from 1 to 8. Then, 108 were used for validation, but confusion matrix points out true positives of up to 16 324. Finally, the subplots where the images were tested seem to be rather limited as well (up to 16 × 10.6 m).
Fan et al. (2018) developed a three step approach for identifying tobacco and non-tobacco plants in VHR RGB image (35 mm). They used morphological operations and watershed segmentation to extract tobacco plants, then a deep convolutional neural network (CNN) is used to classify tobacco plant regions and non-tobacco plants. The authors reported that they could classify tobacco plants with a classification error of less than 4% (Fan et al. 2018). A negative side of this work is that it relies on the fact that central regions of tobacco plants are generally brighter than the leaf regions. Moreover, the images show that result from the application of the morphologic operations and watershed already discriminate the tobacco plants. Because the tobacco plants are in rows and the non-tobacco plants are vegetation outside the row, it is not clear what is the authors’ motivation for using a CNN in this task. Furthermore, the authors’ approach has a high reliance on CNN, which has the downside of needing a lot of training data of multiple species of plants in order to classify all of the plants surrounding the tobacco plants.
While there are several works that address crop vegetation detection (Torres-Sánchez et al. 2015; Li et al. 2019), there are only two works in literature that tackle crop discrimination at a plant level (Li et al. 2019; Fan et al. 2018). Furthermore, there is no automatic approach for plant counting in general wide-row cultivars. The datasets employed in the previous works also make smaller errors and higher metrics rates more attainable due to the selection process or cultivar arrangement. Furthermore, is often reported that the data acquisition is made at different heights, but no sensitivity tests were made for different spatial resolutions. Finally, the approaches proposed were not tested in a different environment conditions or crop growing stages.
The objective of this research is to design a fully automatic algorithm for computing the number of plants in a field using very-high resolution RGB UAV imagery (with a cm pixel resolution, 8–16 mm) to assess the crop emergence. The research question formulated in this work was if it is possible to count the number of plants from very-high resolution RGB UAV imagery. This is done in a VHR RGB ortho-mosaic, build from individual images collected by a UAV (Colomina et al. 2014). The algorithm proposed is a hybrid machine vision-learning approach, because it uses a deep learning (Kamilaris et al. 2018) approach named transfer learning (Kaya et al. 2019) to estimate the average plant area size and direct vegetation segmentation methods for computing the number of crop plants (Hamuda et al. 2016). With transfer learning a neural network model is first trained with general image data. Then, from initially trained models are then used in a new model trained specifically for the problem of interest, i.e. detecting individual plants (Dawei et al. 2019). The output of the algorithm developed will be compared with the two ground truths: UAV images where individual plants have been annotated by hand and the number of seeds planted per linear meter. Due to the nature of this research being one of its kind it does not follow the standard scientific method, but rather a variant of the engineering method (Koen 1985) were in the first place the problem is defined, secondly the data is gathered, then the system is constructed, and finally tested.
The contributions of this work are: (1) the development of an automatic procedure for counting the number of crop plants automatically from VHR UAVs images; (2) description of a methodology for extrapolating verification data from large close-rows crops, and (3) performance analysis of the developed algorithm to different spatial resolutions.
Material and methods
Field test and data acquisition
The experimental area is a spinach field in the province of Flevoland, nearby Lelystad, in the Netherlands. This field has the size of approximately 3.5 ha with a distance of 50 cm between rows. The crop on the field is spinach (Spinacia oleracea) that is approximately 10 weeks old. Figure 1 illustrate a map of the study area. Due to the confidentiality, the field’s exact location and the coordinates have been left out.
A quad-rotor UAV (DJI Phantom 3 PRO, Shenzhen, China) equipped with a digital colour camera (FC300X, Shenzhen, China) was used to survey the spinach field and to acquire RGB imagery. The UAV, aerial mission, and camera specifications are depicted in Tables 1 and 2, respectively.
The aerial surveying flying path was defined using the commercial aerial mission planning software Pix4D capture (Lausanne, Switzerland). The UAV flew autonomously (under supervision of a pilot) over the spinach field at 40 and 20 m height to provide two different VHR spatial resolutions: low and high, respectively. Both datasets have been captured by the UAV on the 21st June of 2018. After the flights the RGB images were transferred to a computer to build the VHR ortho-image using a Agisoft PhotoScan (St. Petersburg, Russia). Two orthorectified images were generated using the parameters shown in Table 3 with a TIF (tagged image file format) extension: 16 mm/pixel and 8 mm/pixel VHR image for the 40 and 20 m flying height, respectively. These different resolutions were achieved by surveying the target area at different heights.
Processing the VHR image
The two VHR images generated in the previous section have a file size of 828 Mb and 670 Mb, for 8 and 16 mm/pixels spatial resolution, respectively. In order to optimize the computational time needed to process large images, a procedure to decompose the images in smaller tiles was applied. The VHR images were tiled into squares to ensure efficient processing and preventing the computer from running out of physical memory. Tile E (10 890 × 9 990 pixels, 87.12 × 79.92 m) will was used to validate the algorithm = due to its uniformity regarding the number of plants on that unit of picture (Fig. 2).
Verification datasets
In order to evaluate the plant count algorithm, the accuracy of the plant count was calculated. This accuracy (Acc, %) was given by:
where the CNP and ANP are the computed number of plants, and approximated number of plants, respectively. Ideally, plant counts obtained from the field are adopted to calculate the accuracy. For this study, the accuracy of the plant count algorithm was evaluated by two verification datasets providing indirect estimations of the number of plants present in the field. Counting based on machine vision approaches potentially has a high accuracy but will not be as accurate as a human interpretation for small areas. In this study, the results obtained with the plant counting algorithm were compared with two data sources: (1) amount of seeds per line of meter, and (2) direct manual plant labelling on the orthomosaic.
According to the field manager who planted the seeds, the field was planted with 8–10 seeds per meter in each row. The distance between rows was 50 cm and each row had an average length of 283 m (Fig. 3). The width of the field was approximately 100 m. Which means there were 200 rows in the whole field and thus in the entire spinach field. With this information, the number of seeds for the whole field was calculated. Each row contained about 2 547 seeds, which means there were 200 × 2 547 = 509 400 seeds planted in the field.
The second verification dataset was defined by manually labelling the number of spinach plants in a small unit of the VHR image. For this purpose, a smaller unit of tile E was cropped (Fig. 3). This area is small enough so that manual counting was feasible and large enough to have enough plants to serve as a sample to perform accuracy tests. Figure 3 illustrates how this sub-unit was obtained from unit E. This unit, also referred as unit X with 1 680 × 1 600 pixels—equivalent to 13.44 × 12.8 m—contains 935 plants.
Once Unit X was fully and manually counted, this information was used to validate the algorithm, but also to estimate the plant number for the other parts of the orthomosaic by extrapolating the results on a larger area. This can be expressed as follows:
where \({Plant\,Number}_{i}\) is the plant number of the unit of image that has to be calculated. \({Area}_{i}\), the area of that same unit of image. \({Plants}_{x}\) are the number of plants in unit X and \({Area}_{x}\) the area of unit x.
Counting crop plants algorithm
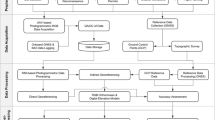
The flowchart of the algorithm for plant counting is shown in Fig. 4. The four-step algorithm has just one input and one output. The red solid line block that addresses plant classification can be either automatic (as shown in Fig. 4) or manual. In the next section the different algorithm blocks will be explained in detail.
Determining the average single plant area
The first step of this approach was to find single spinach plants in the input VHR image that can be used as a reference to estimate the average area of a single plant. To solve this problem, two approaches for computing the average plant area were considered: manual and automatic. The manual approach relies on user annotated single images as inputs (different from the number of plants in field) and the automatic approach employs a deep learning method based on transfer learning. The objective was to train a CNN with single labelled plants and then for each input VHR RGB image (or unit) single plants are identified.
In this case, the AlexNet framework was used as CNN (Krizhevsky et al. 2017). This framework has the advantage that it was already pre-trained with several images and that for the purpose of this study, the main requirement was to have enough data to train the first network layer. For AlexNet to be able to detect individual plants it has to be trained with a number of training samples. In this study AlexNet has been trained to recognize three distinct classes: Individual Plants, Multiple Plants and Background soil (Fig. 5).
The annotation set consisted of 800 manually annotated single plant images, 550 images of multiple plants, and 400 images of background soil. However, 80% of the images were used to train the network and the remaining 20% were used for validation. The training and validation data sets size used are in line with previous works found in literature that used the same training and validation scheme (Song et al. 2014; Giuffrida, et al. 2018).
With the aim to train the AlexNet the input images had a size of 227 × 227 pixels. In the most ideal case to have an orthomosaic in which 227 × 227 pixels correspond to a plant area of approximately 40 × 40 cm, then the orthomosaic spatial resolution had to be approximately 0.7 mm/pixel, which is sub-millimetre level and currently not feasible with commercially available UAVs. Therefore, the images employed for training and validation had to be resized and magnified. Figure 6 shows an example of the original 227 × 227 orthophoto alongside the resized image that is suitable for AlexNet to classify.
After the training of AlexNet, the complete orthomosaic was divided into smaller units (Fig. 2), these units were inputs for AlexNet which then classified plants as an individual plant, multiple plants or simply background. The individual plant images detected using AlexNet were then further processed in order to compute the average number of pixels per plant.
Crop segmentation
The crop segmentation was carried out by applying an Excess Green Index (ExG) to the VHR image and after an Otsu’s threshold, which resulted in a binary image where the true pixels were the discriminated spinach crop. The choice for these algorithms were applied was because of their simplicity, effectiveness, and several reported successful cases (Hamuda et al. 2016). The Excess Green Index (ExG) is a simple algorithm that computes the number of green pixels (vegetation) in an image and can be expressed as:
where r, g and b are the chromatic coordinates derived from:
R′, G′ and B′ are the normalized RGB coordinates ranging from 0 to 1 and can be derived from:
where R, G, B, are the actual pixel values in digital numbers varying between 0 and 255 and Rmax, Gmax and Bmax is the maximum value for the respective colors (255 for a 24-bits images). After, the image was converted to a grey scale to apply the Otsu’s methods. Otsu’s method automatically converts a grey level image into a binary image by performing a clustering based image thresholding (Otsu 1979). The result is a binary bounding plant image where the number of pixels per plant can be obtained and the average single plant area computed.
Counting
The last step of the plant count algorithm was to calculate the number of plants. This was done by the following equation:
where \(I(i,j)\) defines a VHR image with grid pixel at i, j that correspond to a colour vector, and \(\frac{\sum_{k=0}^{n}p(i,j)}{n}\) is the average pixel area of the single plants detected in the orthomosaic.
Results
This section presents the performance of the overall plant counting workflow (Fig. 5). First the performance of AlexNet classification where the goal was to automatically extract a single plant area from an ortho-mosaic unit. Then several tests are carried out applying the overall plant counting workflow: (1) counting plants in a single row image, (2) in the unit X image, and (3) in the complete ortho-mosaic.
AlexNet training and classification results
Table 4 shows the training parameters used for training the AlexNet with 1750 images, the number of iterations, and the time elapsed during the training method. In total, it took 1 min and 7 s to train it with three classes and 150 iterations. These values were achieved with a powerful graphics processing unit GPU (Nvidia GTX 1060 3 GB). Table 4 also shows that after the 50th iteration the mini-batch accuracy reaches 100%.
The overall accuracy is 98.6% (Table 5). Meaning that the model is able to classify the three classes correctly with an accuracy close to that of a human labelling the images (Giuffrida et al. 2018). Table 5 shows the confusion matrix and a table of the Precision, Recall and F1-scores of this test. Precision is the ratio of correctly predicted positive observations to the total predicted positive observations. Recall is the ratio of correctly predicted positive observations to the all observations in actual class. The F1 Score is the weighted average of Precision and Recall. The F1-scores for this training result was 97%, 98% and 97% for the individual plants, background, and multiple plant classes respectively. The F1-score values show that the trained model was suitable for classifying these three classes.
Local counting analysis: row level
In the first test, small units of the image were cut out and processed. The output results were compared to the number of seeds and the actual number of plants counted manually and annotated. In order to do this five sets of five different units of the image were cut out. These images consisted of a single row with different lengths. These images were cut into 1 m, 2 m, 4 m, 5 m, and 10 m lengths. There were in total of 25 images. Figure 7 shows one of each type of image.
Table 6 shows the results obtained when the method is applied to the rows images depicted in Fig. 7. The PC stands for plants counted using the approaches proposed, and Err stands for error between the approaches proposed and the hand counted plants by an expert. The manual and fully automatic methods both yield small errors, a maximum for error of 10% for manual, and 6% for automated. Which means that plants are counted with a maximum error of 3 plants (10 m row image), and 1 to 0.5 plants for the remaining row images.
Unit X counting analysis: patch level
Furthermore, to evaluate the upscaling potential of the proposed approach, these tests were extended to unit X (13.44 × 12.8 m) of the ortho-mosaic (Fig. 3b), considering the whole of unit X was also manually counted by an expert. The algorithm results using manual and automatic approaches are depicted in Table 7 for the 8 mm/pixel ortho-mosaic. To check the sensitivity of the algorithm the same algorithm with the same methods were applied to a secondary dataset with a different spatial resolution (16 mm/pixel). The results of this second run can be seen in Table 8.
The accuracy results were very much in line with the calculations of the single rows (Table 6). In this case, the area was much larger compared to the single row image and 40 plants where overestimated in a 172 m2 area with the automated approach that performed better. The error in both spatial resolutions using the automated approach is not greater than 5%, and with a minimum error difference between approaches from 0.7%. Nevertheless, that difference improves counting accuracy by about 6 plants.
Assessing the amount of plants in the complete orthomosaics
The number of plants in the complete orthomosaic was estimated by applying the proposed approach to each orthomosaic unit and compared with an extrapolated number of plants. The last was obtained by computing the extrapolated number of plants (ENP) for each orthomosaic unit. The computed number of plants for each orthomosaic is shown in Fig. 8 for spatial resolution 8 mm/pixel and 16 mm/pixel. These results are also presented for the manual single plants annotated approach and automatic approach using AlexNet.
There are two important things to be noticed: first, the manual values are higher than the automatic count, and second, there is an overestimation of number of plants regarding the ENP. The same overestimation was also noticed in previous analysis at the row level and patch level for the 8 mm/pixel orthomosaic, but this overestimation was less noticed due to the small size of the images processed. Figure 9 shows the average error using manual and automatic approaches and respective spatial resolutions. The results show that the error was smaller when using the 8 mm/pixel orthomosaic and when using the automatic approach. The maximum difference between manual and automatic approach is 9.4% (unit D—8 mm/pixel) and 6.2% (unit G—16 mm/pixel).
Although the error presented in Fig. 9 looks high when comparing it with the previous test at the row level and patch level, should be noticed that these errors are due to a propagation error that is expected to grow with the size of the areas. If the total number of plants in the orthomosaic is computed with the propagation error equation that is below,
where \({e}_{i}\) is the error of the i-th unit of the orthomosaic given by,
In that case, the errors obtained for each orthomosaic (spatial resolution 8 mm/pixel and 16 mm/pixel) would also be from about 42.5% and 50% respectively.
Performance over imagery acquired in a previous year campaign (2017)
Finally, to analyse how the algorithm would behave in a complete different—spatial resolution, crop stage, and environment conditions—image dataset, another evaluation was carried out where plants were hand counted in situ in an area of 9.3 m2. In this experiment two spatial resolutions where considered: 4 mm/pixel and 18 mm/pixel. Inside the yellow tape square depicted in Fig. 10, 32 plants where counted. With the proposed automatic algorithm, the number of plants computed for the 4 mm/pixel unit was 31 plants, and 18 mm/pixel was 35 plants. This experiment once again showed that the algorithm is suitable for estimating the number of plants in the field. These results are within the previous errors because the maximum error that could be obtained was less than 9%.
Discussion
The outcomes from this study demonstrated that the was possible to automatically compute the number of plants using this approach at a row level (up to 5 linear meters) with an average error of 3.4%, at a patch level (up to 172 m2) with an error of 5%. However, the study demonstrated as well that automatically computing the number of plants at the level of the orthomosaic units (up to 3 259 m2) resulted in an error between 25 and 72%, and for the complete orthomosaic of 35 000 m2 with an error of 42.5%, which is a very high error rate. Reasons for this are explained below.
A very important metric in studies about yield estimation and machine vision is the extrapolated plant number. This information is crucial for validation purposes. In this study getting good plant number information was not a trivial task given the field size. Image labelling is not only very time consuming and resource demanding but often also subject to human error (Ghosal et al. 2018; Sa et al. 2017). Just manually counting a small unit of image with about 950 plants took about 3 h of manual work (Fig. 4). Counting 8 unit of images with at least 20 000 plants would be time intensive while manual counting also can include errors.
The direct methods as elaborated in this paper were employed following the study of Hamuda et al. (2016), in which the ExtG was successfully used in many precision agriculture applications. Deep learning was successfully employed in the past for counting in agriculture. For example, Qureshi et al. (2016) used this approach to detect mango fruits on trees using a handheld camera. The previous work from Fan et al. 2018, addressing crop emergence using deep learning could not be used to address the problem addressed in this study, because it assumed that the tobacco plant crown has a lighter crown colour, which is not this case, and because classification aims are different. In the proposed approach the classification aim is to get individual plants and calculate the average number of pixels at the plant level. False positives can affect the calculated average size of a plant, if the false positives are significantly larger than a single plant, the average size will also increase. But from the test results and the F1-scores, it is possible to conclude that the chance of this occurring is low (Table 5).
While seeds planted by the land owners gave an estimate on how many plants potentially could have emerged, it is not accurate enough to employ as a ground truth. Because the crop emergence is the outcome of the proposed approach divided by the number of seeds planted. The number of seeds is a very high over-estimation of the number of actual plants that have successfully germinated and thus actually became plants. The crop emergence in unit X (Tables 7, 8) was only 30%. The number of seeds is nearly three times the number of plants that have been counted. These figures were afterwards discussed with the field owner which pointed out that these values match with poor crop emergence during that season. There where large areas where seeds did not emerge.
The extrapolation of the number of seeds and plants from a small unit of land was used to be able to provide a plant number reference for the entire orthomosaic. This strategy was assumed reliable because the correlation between the seeds on the field and extrapolated plant number per orthomosaic unit was found high (Figs. 8, 9). The final accuracy error for the entire orthomosaic was found very high based on the extrapolated plant number per orthomosaic unit. This was explained by the propagation error through the orthomosaic unit that cover larger areas with more plants than single rows and unit X patch.
The results obtained at the row level with several lengths (Table 6), at the patch level (small area) in unit X (Table 7), and another dataset from another year showed that the algorithm performed with a maximum error of 10%. The maximum errors achieved with the proposed approach were lower than the errors reported in similar studies where other image sensors where employed (Tyystjärvi et. al. 2011). Previous work in plant counting extended the analysis to different spatial resolutions (Li et al. 2019) and alternative object localization approaches (Ribera et al. 2019). The approach proposed was tested in two different image spatial resolutions. The results indicated that the maximum error obtained when decreasing the spatial resolution by a factor of 2 is from 0.7% which correspond to 6 plants (see Tables 7, 8) at a patch level, and 15% for an orthomosaic unit which correspond to a difference of 1 977 plants in 2 695 m2 area.
In this study, the counting algorithm that was used is a very simple approach that has some limitations. First, it assumes that all the plants in the field are spinach. It is inevitable that some unknown species of weeds can be in between the rows. Moreover, the actual formula used to calculate the number of plants by dividing the number of plant pixels by the average number of pixels per plant will not distinguish between closely growing plants. When two plants grow very close to each other there is bound to be overlap between the plants. This results in fewer plant pixels in the binary image which in turn results into a lower number of total plant pixels in the image. This incorrectly results in a lower number of plants counted than there should be. This type of close vegetation also results in a difficulty counting for a human being as the overlap makes it hard to tell if a plant is one plant with a large canopy or two very closely growing plant. In this study, the image has been processed by dividing it into small chunks of 50 × 50 pixels and 25 × 25 pixels for the 8 mm and the 16 mm resolution respectively. Afterward, each of these chunks has been resized to the appropriate input size. This approach was developed and tested on very high-resolution RGB UAV imagery with spatial resolutions from 4 mm/pixel to 18 mm/pixel, but it would not work for lower resolution imagery due to the fact these input images would lose too much detail and become too blurry.
Conclusions
This research has presented a design for a plant counting method in which machine vision and transfer learning using AlexNet are combined to address the problem of detecting from a very high-resolution RGB UAV image the number of plants on the field that emerged after sowing.
While there were previously some studies that addressed crop vegetation segmentation to estimate yield, there were only a few that addressed how to count plants from very high-resolution RGB UAV images. these studies did not provide enough detail about performance of the methodologies when subject to heterogeneous datasets. Also they are less likely to work in other crops if they depend on crop specific colour features.
The full-automatic method presented in this study computed a total number of 170 000 plants in an area of 3.5 ha with an error of 42.5%. This means that the algorithm estimated the numbers of plants in a wide-row crop field with an accuracy of 67.5%. It was shown that up to 44% of the seeds grew into plants.
The algorithm was tested on 8 mm and 16 mm per pixel spatial resolutions and estimated the number of plants with a maximum difference of 15%. However, the single plant classification performance decreased significantly if the spatial resolution was smaller. This was due to the limitation of AlexNet only being able to use images of 227 by 227 pixels. While the algorithm still performed well with 16 mm, this might not be the case for a lower resolution. For that reason future research will be to employ a convolutional neural network for training and classification of single plants.
References
Colomina, P. M. (2014). Unmanned aerial systems for photogrammetry and remote sensing: A review. ISPRS Journal of Photogrammetry and Remote Sensing, 92, 79–97. https://doi.org/10.1016/j.isprsjprs.2014.02.013.
Dawei, W., Limiao, D., Jiangong, N., Jiyue, G., Hongfei, Z., & Zhongzhi, H. (2019). Recognition pest by image-based transfer learning. Journal of the Science of Food and Agriculture, 99, 4524–4531. https://doi.org/10.1002/jsfa.9689.
Fan, Z., Lu, J., Gong, M., Xie, H., & Goodman, E. D. (2018). Automatic Tobacco Plant Detection in UAV images via deep neural networks. IEEE Journal of Selected Topics in Applied Earth Observations and Remote Sensing, 11(3), 876–887. https://doi.org/10.1109/jstars.2018.2793849.
Giuffrida, M. V., Doerner, P., & Tsaftaris, S. A. (2018). Pheno-Deep Counter: A unified and versatile deep learning architecture for leaf counting. Plant Journal, 96, 880–890.
Ghosal, S., Blystone, D., Singh, A., Ganapathysubramanian, B., Singh, A., & Sarkar, S. (2018). An explainable deep machine vision framework for plant stress phenotyping. Proceedings of the National Academy of Sciences United States of America, 11(18), 4613–4618. https://doi.org/10.1073/pnas.1716999115.
Guo, W., Zheng, B., Potgieter, A. B., Diot, J., Watanabe, K., Noshita, K., et al. (2018). Aerial imagery analysis: Quantifying appearance and number of sorghum heads for applications in breeding and agronomy. Frontiers in Plant Science, 9, 1544. https://doi.org/10.3389/fpls.2018.01544.
Hamuda, E., Glavin, M., & Jones, E. (2016). A survey of image processing techniques for plant extraction and segmentation in the field. Computers and Electronics in Agriculture, 125, 184–199. https://doi.org/10.1016/j.compag.2016.04.024.
Hayes, M. J., & Decker, W. L. (1996). Using NOAA AVHRR data to estimate maize production in the United States Corn Belt. International Journal of Remote Sensing, 17(16), 3189–3200. https://doi.org/10.1080/01431169608949138.
Hunt, E. R., Hively, W. D., Fujikawa, S., Linden, D., Daughtry, C. S., McCarty, G., et al. (2010). Acquisition of NIR-green-blue digital photographs from unmanned aircraft for crop monitoring. Remote Sensing, 2(1), 290–305. https://doi.org/10.3390/rs2010290.
Kamilaris, A., & Prenafeta-Boldú, F. X. (2018). Deep learning in agriculture: A survey. Computers and Electronics in Agriculture, 147, 70–90. https://doi.org/10.1016/j.compag.2018.02.016.
Kaya, A., Keceli, A. S., Catal, C., Yalic, H. Y., Temucin, H., & Tekinerdogan, B. (2019). Analysis of transfer learning for deep neural network based plant classification models. Computers and Electronics in Agriculture, 158, 20–29. https://doi.org/10.1016/j.compag.2019.01.041.
Koen, B. V. (1985). Definition of the engineering method. Washington, DC: ASEE Publications.
Krizhevsky, A., Sutskever, I., & Hinton, G. H. (2017). ImageNet classification with deep convolutional neural networks. Communications of the ACM, 60(6), 84–90. https://doi.org/10.1145/3065386.
Li, B., Xu, X., Han, J., Zhang, L., Bian, C., Jin, L., et al. (2019). The estimation of crop emergence in potatoes by UAV RGB imagery. Plant Methods. https://doi.org/10.1186/s13007-019-0399-7.
Montalvo, M., Pajares, G., Guerrero, J. M., Romeo, J., Guijarro, M., Ribeiro, A., et al. (2012). Automatic detection of crop rows in maize fields with high weeds pressure. Expert Systems with Applications. https://doi.org/10.1016/j.eswa.2012.02.117.
Otsu, N. (1979). A threshold selection method from gray-level histograms. IEEE Transactions on Systems, Man, and Cybernetics, 9(1), 62–66. https://doi.org/10.1109/TSMC.1979.4310076.
Qureshi, W. S., Payne, A., Walsh, K. B., Linker, R., Cohen, O., & Dailey, M. N. (2016). Machine vision for counting fruit on mango tree canopies. Precision Agriculture, 18, 224–244.
Reza, M. N., Na, I. S., Baek, S. W., & Lee, K. H. (2019). Rice yield estimation based on K-means clustering with graph-cut segmentation using low-altitude UAV images. Biosystems Engineering, 177, 109–121. https://doi.org/10.1016/j.biosystemseng.2018.09.014.
Ribera, J., Güera, D., Chen, Y., & Delp, E. J. (2019). Locating objects without bounding boxes. In 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, pp. 6472–6482.
Rokhmana, C. A. (2015). The potential of UAV-based remote sensing for supporting precision agriculture in Indonesia. Procedia Environmental Sciences, 24, 245–253. https://doi.org/10.1016/J.PROENV.2015.03.032.
Sarron, J., Malézieux, É., Sané, C., Faye, É., Sarron, J., Malézieux, É., et al. (2018). Mango yield mapping at the orchard scale based on tree structure and land cover assessed by UAV. Remote Sensing, 10(12), 1900. https://doi.org/10.3390/rs10121900.
Senthilnath, J., Dokania, A., Kandukuri, M., Anand, G., & Omkar, S. (2016). Detection of tomatoes using spectral-spatial methods in remotely sensed RGB images captured by UAV. Biosystems Engineering. https://doi.org/10.1016/j.biosystemseng.2015.12.003.
Sa, I., Chen, Z., Popovic, M., Khanna, R., Liebisch, F., Nieto, J., et al. (2017). weedNet: dense semantic weed classification using multispectral images and MAV for smart farming. IEEE Robotics and Automation Letters, 3, 588–595. https://doi.org/10.1109/LRA.2017.2774979.
Som-ard, J., Hossain, M. D., Ninsawat, S., & Veerachitt, V. (2018). Pre-harvest sugarcane yield estimation using UAV-based RGB images and ground observation. Sugar Tech, 20, 645–657. https://doi.org/10.1007/s12355-018-0601-7.
Song, Y., Glasbey, C. A., Horgan, G. W., Polder, G., Dieleman, J. A., & van der Heijden, G. W. A. M. (2014). Automatic fruit recognition and counting from multiple images. Biosystems Engineering, 118, 203–215.
Tokekar, P., Hook, J. V., Mulla, D., & Isler, V. (2016). Sensor planning for a symbiotic UAV and UGV system for precision agriculture. IEEE Transactions on Robotics, 32(6), 1498–1511. https://doi.org/10.1109/TRO.2016.2603528.
Tyystjärvi, E., Nørremark, M., Mattila, H., Keränen, M., Hakala-Yatkin, M., Ottosen, C.-O., et al. (2011). Automatic identification of crop and weed species with chlorophyll fluorescence induction curves. Precision Agriculture., 12, 546–563. https://doi.org/10.1007/s11119-010-9201-6.
Torres-Sánchez, J., López-Granados, F., & Peña, J. M. (2015). An automatic object-based method for optimal thresholding in UAV images: Application for vegetation detection in herbaceous crops. Computers and Electronics in Agriculture, 114, 43–52. https://doi.org/10.1016/J.COMPAG.2015.03.019.
Yeom, J., Jung, J., Chang, A., Maeda, M., & Landivar, J. (2018). Automated open cotton boll detection for yield estimation using unmanned aircraft vehicle (UAV) Data. Remote Sensing, 10, 1895.
Acknowledgements
This work was supported by the SPECTORS project (143081) which is funded by the European cooperation program INTERREG Deutschland-Nederland.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Valente, J., Sari, B., Kooistra, L. et al. Automated crop plant counting from very high-resolution aerial imagery. Precision Agric 21, 1366–1384 (2020). https://doi.org/10.1007/s11119-020-09725-3
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11119-020-09725-3