Abstract
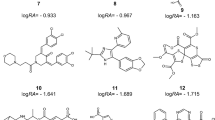
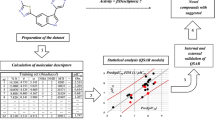
Based on the mechanism of action of PKC-θ, the inhibition of this enzyme is considered a potential target for the treatment of autoimmune diseases such as rheumatoid arthritis (RA), inflammatory bowel disease (IBD), and psoriasis. In the present study, 57 structurally diverse tricyclic triazinone analogues as potential PKC-θ inhibitors has been taken into consideration for QSAR analysis through Monte Carlo optimization. QSAR models are developed using the balance of correlation method in the CORAL software with two target functions (TF1 and TF2). The models constructed with IIC are found more robust and authentic. The established QSAR model with best \( {R}_{\mathrm{calibration}}^2 \) = 0.9653 for split 3 is considered the topmost model. The predictabilities of the recommended QSAR model are assessed through various statistical parameters. The outlier of each model is also identified using the applicability domain (AD). The common mechanistic interpretation of the increasing and decreasing attributes has been extracted by evaluating the correlation weights of diverse structural attributes obtained in three Monte Carlo optimization runs from two splits, i.e., split 3 and 4. In the last, the result of mechanistic interpretation is validated by performing the docking studies of compounds PKC03, PKC07, PKC16, PKC25, and PKC56 in the experimental structure of protein kinase C-θ (PDB ID: 4Q9Z). The numerical value of the correlation coefficient between calculated activity and binding affinity is found 0.9597. Hence, the developed QSAR models are descriptive and predictive in nature and the results are in sound agreement with the experimental observations.

Graphical abstract


Similar content being viewed by others
References
Zanin-Zhorov A, Kumari S, Hippen KL, Merkel SC, MacMillan ML, Blazar BR, Dustin ML (2017) Human in vitro-induced regulatory T cells display Dlgh1dependent and PKC-theta restrained suppressive activity. Sci Rep 7(1):4258. https://doi.org/10.1038/s41598-017-04053-5
Han C, Lei D, Liu L, Xie S, He L, Wen S, Zhou H, Ma T, Li S (2020) Morphine induces the differentiation of T helper cells to Th2 effector cells via the PKC-theta-GATA3 pathway. Int Immunopharmacol 80:106133. https://doi.org/10.1016/j.intimp.2019.106133
von Essen MR, Kongsbak M, Levring TB, Hansen AK, Boding L, Lauritsen JP, Woetmann A, Baier G, Odum N, Bonefeld CM, Geisler C (2013) PKC-theta exists in an oxidized inactive form in naive human T cells. Eur J Immunol 43(6):1659–1666. https://doi.org/10.1002/eji.201243140
Wachowicz K, Baier G (2014) Protein kinase Ctheta: the pleiotropic T-cell signalling intermediate. Biochem Soc Trans 42(6):1512–1518. https://doi.org/10.1042/BST20140179
Cole DC, Asselin M, Brennan A, Czerwinski R, Ellingboe JW, Fitz L, Greco R, Huang X, Joseph-McCarthy D, Kelly MF, Kirisits M, Lee J, Li Y, Morgan P, Stock JR, Tsao DH, Wissner A, Yang X, Chaudhary D (2008) Identification, characterization and initial hit-to-lead optimization of a series of 4-arylamino-3-pyridinecarbonitrile as protein kinase C theta (PKCtheta) inhibitors. J Med Chem 51(19):5958–5963. https://doi.org/10.1021/jm800214a
Sakowicz-Burkiewicz M, Nishanth G, Helmuth U, Drogemuller K, Busch DH, Utermohlen O, Naumann M, Deckert M, Schluter D (2008) Protein kinase C-theta critically regulates the proliferation and survival of pathogen-specific T cells in murine listeriosis. J Immunol 180(8):5601–5612. https://doi.org/10.4049/jimmunol.180.8.5601
Solomou EE, Juang YT, Tsokos GC (2001) Protein kinase C-theta participates in the activation of cyclic AMP-responsive element-binding protein and its subsequent binding to the -180 site of the IL-2 promoter in normal human T lymphocytes. J Immunol 166(9):5665–5674. https://doi.org/10.4049/jimmunol.166.9.5665
Thuille N, Siegmund K, Klepsch V, Schorgenhuber J, Danklmaier S, Leitges M, Baier G (2019) Loss-of-function phenotype of a PKCtheta(T219A) knockin mouse strain. Cell Commun Signal 17(1):141. https://doi.org/10.1186/s12964-019-0466-8
Cywin CL, Dahmann G, Prokopowicz 3rd AS, Young ER, Magolda RL, Cardozo MG, Cogan DA, Disalvo D, Ginn JD, Kashem MA, Wolak JP, Homon CA, Farrell TM, Grbic H, Hu H, Kaplita PV, Liu LH, Spero DM, Jeanfavre DD, O'Shea KM, White DM, Woska Jr JR, Brown ML (2007) Discovery of potent and selective PKC-theta inhibitors. Bioorg Med Chem Lett 17(1):225–230. https://doi.org/10.1016/j.bmcl.2006.09.056
Tanaka Y, Altman A (2002) T cell signaling: protein kinase Cθ the immunological synapse and characterization of SLAT a novel T helper 2-specific adapter protein. Allergol Int 51 (3):167–174. https://doi.org/10.1046/j.1440-1592.2002.00261.x
Niu C, Boschelli DH, Tumey LN, Bhagirath N, Subrath J, Shim J, Wang Y, Wu B, Eid C, Lee J, Yang X, Brennan A, Chaudhary D (2009) First generation 5-vinyl-3-pyridinecarbonitrile PKCtheta inhibitors. Bioorg Med Chem Lett 19(20):5829–5832. https://doi.org/10.1016/j.bmcl.2009.08.086
Salek-Ardakani S, So T, Halteman BS, Altman A, Croft M (2004) Differential regulation of Th2 and Th1 lung inflammatory responses by protein kinase C theta. J Immunol 173(10):6440–6447. https://doi.org/10.4049/jimmunol.173.10.6440
Healy AM, Izmailova E, Fitzgerald M, Walker R, Hattersley M, Silva M, Siebert E, Terkelsen J, Picarella D, Pickard MD, LeClair B, Chandra S, Jaffee B (2006) PKC-theta-deficient mice are protected from Th1-dependent antigen-induced arthritis. J Immunol 177(3):1886–1893. https://doi.org/10.4049/jimmunol.177.3.1886
Salek-Ardakani S, So T, Halteman BS, Altman A, Croft M (2005) Protein kinase Ctheta controls Th1 cells in experimental autoimmune encephalomyelitis. J Immunol 175(11):7635–7641. https://doi.org/10.4049/jimmunol.175.11.7635
Tan SL, Zhao J, Bi C, Chen XC, Hepburn DL, Wang J, Sedgwick JD, Chintalacharuvu SR, Na S (2006) Resistance to experimental autoimmune encephalomyelitis and impaired IL-17 production in protein kinase C theta-deficient mice. J Immunol 176(5):2872–2879. https://doi.org/10.4049/jimmunol.176.5.2872
Nagahama K, Ogawa A, Shirane K, Shimomura Y, Sugimoto K, Mizoguchi A (2008) Protein kinase C theta plays a fundamental role in different types of chronic colitis. Gastroenterology 134(2):459–469. https://doi.org/10.1053/j.gastro.2007.11.005
Dushin RG, Nittoli T, Ingalls C, Boschelli DH, Cole DC, Wissner A, Lee J, Yang X, Morgan P, Brennan A, Chaudhary D (2009) Synthesis and PKCtheta inhibitory activity of a series of 4-indolylamino-5-phenyl-3-pyridinecarbonitriles. Bioorg Med Chem Lett 19(9):2461–2463. https://doi.org/10.1016/j.bmcl.2009.03.053
Tumey LN, Bhagirath N, Brennan A, Brooijmans N, Lee J, Yang X, Boschelli DH (2009) 5-Vinyl-3-pyridinecarbonitrile inhibitors of PKCtheta: optimization of enzymatic and functional activity. Bioorg Med Chem 17(23):7933–7948. https://doi.org/10.1016/j.bmc.2009.10.020
Basak SC, Vracko MG (2020) Parsimony principle and its proper use/ application in computer-assisted drug design and QSAR. Curr Comput Aided Drug Des 16(1):1–5. https://doi.org/10.2174/157340991601200106122854
Veeravarapu H, Malkhed V, Mustyala KK, Vadija R, Malikanti R, Vuruputuri U, Muthyala MKK (2020) Structure-based drug design, synthesis and screening of MmaA1 inhibitors as novel anti-TB agents. Mol Divers. https://doi.org/10.1007/s11030-020-10107-0
Senthil R, Sakthivel M, Usha S (2020) Structure-based drug design of peroxisome proliferator-activated receptor gamma inhibitors: ferulic acid and derivatives. J Biomol Struct Dyn:On Line Published. https://doi.org/10.1080/07391102.2020.1740790
Sadhasivam A, Nagarajan H, Umashankar V (2020) Structure-based drug target prioritisation and rational drug design for targeting Chlamydia trachomatis eye infections. J Biomol Struct Dyn 38(11):3131–3143. https://doi.org/10.1080/07391102.2019.1652691
Kumar P, Kumar A (2020) CORAL: QSAR models of CB1 cannabinoid receptor inhibitors based on local and global SMILES attributes with the index of ideality of correlation and the correlation contradiction index. Chemometr Intel Lab Syst 200:103982. https://doi.org/10.1016/j.chemolab.2020.103982
Kumar A, Sindhu J, Kumar P (2020) In-silico identification of fingerprint of pyrazolyl sulfonamide responsible for inhibition of N-myristoyltransferase using Monte Carlo method with index of ideality of correlation. J Biomol Struct Dyn:On Line Published. https://doi.org/10.1080/07391102.2020.1784286
Puratchikody A, Prabu SL, Umamaheswari A (2019) Computer applications in drug discovery and development. Advances in medical technologies and clinical practice (AMTCP) book series. Medical Information Science Reference, Hershey, PA
Worachartcheewan A, Mandi P, Prachayasittikul V, Toropova AP, Toropov AA, Nantasenamat C (2014) Large-scale QSAR study of aromatase inhibitors using SMILES-based descriptors. Chemometr Intel Lab Syst 138:120–126. https://doi.org/10.1016/j.chemolab.2014.07.017
Doucet J-P, Panaye A (2010) Three dimensional QSAR : applications in pharmacology and toxicology. QSAR in environmental and health sciences. CRC Press, Boca Raton
Qi R, Pan Y, Cao J, Jia Z, Jiang J (2020) The cytotoxicity of nanomaterials: modeling multiple human cells uptake of functionalized magneto-fluorescent nanoparticles via nano-QSAR. Chemosphere 249:126175. https://doi.org/10.1016/j.chemosphere.2020.126175
Mondal D, Ghosh K, Baidya ATK, Gantait AM, Gayen S (2020) Identification of structural fingerprints for in vivo toxicity by using Monte Carlo based QSTR modeling of nitroaromatics. Toxicol Mechan Methods 30 (4):257–265. https://doi.org/10.1080/15376516.2019.1709238
Jain S, Bhardwaj B, Amin SA, Adhikari N, Jha T, Gayen S (2020) Exploration of good and bad structural fingerprints for inhibition of indoleamine-2,3-dioxygenase enzyme in cancer immunotherapy using Monte Carlo optimization and Bayesian classification QSAR modeling. J Biomol Struct Dyn 38 (6):1683–1696. https://doi.org/10.1080/07391102.2019.1615000
Ahmadi S, Ghanbari H, Lotfi S, Azimi N (2020) Predictive QSAR modeling for the antioxidant activity of natural compounds derivatives based on Monte Carlo method. Mol Divers. https://doi.org/10.1007/s11030-019-10026-9
Toropov AA, Toropova AP, Selvestrel G, Benfenati E (2019) Idealization of correlations between optimal simplified molecular input-line entry system-based descriptors and skin sensitization. SAR QSAR Environ Res 30(6):447–455. https://doi.org/10.1080/1062936X.2019.1615547
Toropova MA, Veselinovic AM, Veselinovic JB, Stojanovic DB, Toropov AA (2015) QSAR modeling of the antimicrobial activity of peptides as a mathematical function of a sequence of amino acids. Comput Biol Chem 59 Pt A:126-130. https://doi.org/10.1016/j.compbiolchem.2015.09.009
Toropova AP, Schultz TW, Toropov AA (2016) Building up a QSAR model for toxicity toward Tetrahymena pyriformis by the Monte Carlo method: a case of benzene derivatives. Environ Toxicol Pharmacol 42:135–145. https://doi.org/10.1016/j.etap.2016.01.010
Toropova AP, Toropov AA, Veselinovic JB, Miljkovic FN, Veselinovic AM (2014) QSAR models for HEPT derivates as NNRTI inhibitors based on Monte Carlo method. Eur J Med Chem 77:298–305. https://doi.org/10.1016/j.ejmech.2014.03.013
Ibezim E, Duchowicz PR, Ortiz EV, Castro EA (2012) QSAR on aryl-piperazine derivatives with activity on malaria. Chemometr Intel Lab Syst 110(1):81–88. https://doi.org/10.1016/j.chemolab.2011.10.002
Kumar A, Manisha, Bagri K, Kumar P (2020) Use of graph based descriptors for determination of structural features causing modulation of fructose-1,6-bisphosphatase. Drug Res (Stuttg) 70 (5):226–232. https://doi.org/10.1055/a-1138-8725
Kumar P, Kumar A, Sindhu J (2019) Design and development of novel focal adhesion kinase (FAK) inhibitors using Monte Carlo method with index of ideality of correlation to validate QSAR. SAR QSAR Environ Res 30 (2):63–80. https://doi.org/10.1080/1062936X.2018.1564067
Kumar P, Kumar A, Sindhu J (2019) In silico design of diacylglycerol acyltransferase-1 (DGAT1) inhibitors based on SMILES descriptors using Monte-Carlo method. SAR QSAR Environ Res 30(8):525–541. https://doi.org/10.1080/1062936X.2019.1629998
Toropova AP, Toropov AA, Carnesecchi E, Benfenati E, Dorne JL (2020) The index of ideality of correlation: models for flammability of binary liquid mixtures. Chem Pap 74(2):601–609. https://doi.org/10.1007/s11696-019-00903-w
Kumar A, Kumar P (2020) Construction of pioneering quantitative structure activity relationship screening models for abuse potential of designer drugs using index of ideality of correlation in Monte Carlo optimization. Arch Toxicol 94 (9):3069–3086. https://doi.org/10.1007/s00204-020-02828-w
Ahmadi S (2020) Mathematical modeling of cytotoxicity of metal oxide nanoparticles using the index of ideality correlation criteria. Chemosphere 242:125192. https://doi.org/10.1016/j.chemosphere.2019.125192
Kumar P, Kumar A, Sindhu J, Lal S (2019) QSAR models for nitrogen containing monophosphonate and bisphosphonate derivatives as human farnesyl pyrophosphate synthase inhibitors based on Monte Carlo method. Drug Res (Stuttg) 69 (3):159–167. https://doi.org/10.1055/a-0652-5290
Achary PGR, Toropova AP, Toropov AA (2019) Combinations of graph invariants and attributes of simplified molecular input-line entry system (SMILES) to build up models for sweetness. Food Res Int 122:40–46. https://doi.org/10.1016/j.foodres.2019.03.067
Meng L, Feng K, Ren Y (2018) Molecular modelling studies of tricyclic triazinone analogues as potential PKC-θ inhibitors through combined QSAR, molecular docking and molecular dynamics simulations techniques. J Taiwan Inst Chem Eng 91:155–175. https://doi.org/10.1016/j.jtice.2018.06.017
George DM, Breinlinger EC, Argiriadi MA, Zhang Y, Wang J, Bansal-Pakala P, Duignan DB, Honore P, Lang Q, Mittelstadt S, Rundell L, Schwartz A, Sun J, Edmunds JJ (2015) Optimized protein kinase Ctheta (PKCtheta) inhibitors reveal only modest anti-inflammatory efficacy in a rodent model of arthritis. J Med Chem 58(1):333–346. https://doi.org/10.1021/jm5013006
George DM, Breinlinger EC, Friedman M, Zhang Y, Wang J, Argiriadi M, Bansal-Pakala P, Barth M, Duignan DB, Honore P, Lang Q, Mittelstadt S, Potin D, Rundell L, Edmunds JJ (2015) Discovery of selective and orally bioavailable protein kinase C theta (PKCtheta) inhibitors from a fragment hit. J Med Chem 58(1):222–236. https://doi.org/10.1021/jm500669m
Marvin-Sketch-v.14.11.17.0 (2014). ChemAxon, XhemAxon KFT. Budapest, Hungary
O'Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR (2011) Open babel: an open chemical toolbox. J Cheminform 3 (1):33. https://doi.org/10.1186/1758-2946-3-33
Veselinovic JB, Nikolic GM, Trutic NV, Zivkovic JV, Veselinovic AM (2015) Monte Carlo QSAR models for predicting organophosphate inhibition of acetycholinesterase. SAR QSAR Environ Res 26(6):449–460. https://doi.org/10.1080/1062936X.2015.1049665
Gissi A, Toropov AA, Toropova AP, Nicolotti O, Carotti A, Benfenati E (2014) Building up QSAR model for toxicity of psychotropic drugs by the Monte Carlo method. Struct Chem 25(4):1067–1073. https://doi.org/10.1007/s11224-013-0380-4
Kumar A, Chauhan S (2017) Monte Carlo method based QSAR modelling of natural lipase inhibitors using hybrid optimal descriptors. SAR QSAR Environ Res 28(3):179–197. https://doi.org/10.1080/1062936X.2017.1293729
Manisha CS, Kumar P, Kumar A (2019) Development of prediction model for fructose- 1,6- bisphosphatase inhibitors using the Monte Carlo method. SAR QSAR Environ Res 30(3):145–159. https://doi.org/10.1080/1062936X.2019.1568299
Toropova AP, Toropov AA (2019) QSPR and nano-QSPR: what is the difference? J Mol Struct 1182:141–149. https://doi.org/10.1016/j.molstruc.2019.01.040
Toropov AA, Toropova AP (2017) The index of ideality of correlation: a criterion of predictive potential of QSPR/QSAR models? Mutat Res Genet Toxicol Environ Mutagen 819:31–37. https://doi.org/10.1016/j.mrgentox.2017.05.008
Toropov AA, Toropova AP (2018) Predicting cytotoxicity of 2-Phenylindole derivatives against breast cancer cells using index of ideality of correlation. Anticancer Res 38(11):6189–6194. https://doi.org/10.21873/anticanres.12972
Environment-directorate guidance document on the validation of (quantitative) structure-activity relationship [(Q)SAR] models. Organisation for Economic Co-operation and Development. http://www.oecd.org/officialdocuments/publicdisplaydocumentpdf/?doclanguage=en&cote=env/jm/mono(2007)2. Accessed 07 July 2020
Aher RB, Roy K (2017) Exploring the structural requirements in multiple chemical scaffolds for the selective inhibition of Plasmodium falciparum calcium-dependent protein kinase-1 (PfCDPK-1) by 3D-pharmacophore modelling, and docking studies. SAR QSAR Environ Res 28 (5):390–414. https://doi.org/10.1080/1062936X.2017.1326401
Bhayye SS, Roy K, Saha A (2016) Pharmacophore generation, atom-based 3D-QSAR, HQSAR and activity cliff analyses of benzothiazine and deazaxanthine derivatives as dual A2A antagonists/MAOB inhibitors. SAR QSAR Environ Res 27(3):183–202. https://doi.org/10.1080/1062936X.2015.1136840
Golbraikh A, Tropsha A (2002) Beware of q2! J Mol Graph Model 20(4):269–276. https://doi.org/10.1016/s1093-3263(01)00123-1
Shi LM, Fang H, Tong W, Wu J, Perkins R, Blair RM, Branham WS, Dial SL, Moland CL, Sheehan DM (2001) QSAR models using a large diverse set of estrogens. J Chem Inf Comput Sci 41(1):186–195. https://doi.org/10.1021/ci000066d
Schuurmann G, Ebert RU, Chen J, Wang B, Kuhne R (2008) External validation and prediction employing the predictive squared correlation coefficient test set activity mean vs training set activity mean. J Chem Inf Model 48(11):2140–2145. https://doi.org/10.1021/ci800253u
Pratim Roy P, Paul S, Mitra I, Roy K (2009) On two novel parameters for validation of predictive QSAR models. Molecules 14(5):1660–1701. https://doi.org/10.3390/molecules14051660
Roy K, Chakraborty P, Mitra I, Ojha PK, Kar S, Das RN (2013) Some case studies on application of “r(m)2” metrics for judging quality of quantitative structure-activity relationship predictions: emphasis on scaling of response data. J Comput Chem 34 (12):1071–1082. https://doi.org/10.1002/jcc.23231
Gramatica P, Sangion A (2016) A historical excursus on the statistical validation parameters for QSAR models: a clarification concerning metrics and terminology. J Chem Inf Model 56 (6):1127–1131. https://doi.org/10.1021/acs.jcim.6b00088
Toropova AP, Toropov AA (2019) The index of ideality of correlation: improvement of models for toxicity to algae. Nat Prod Res 33(15):2200–2207. https://doi.org/10.1080/14786419.2018.1493591
Toropov AA, Toropova AP (2019) Use of the index of ideality of correlation to improve predictive potential for biochemical endpoints. Toxicol Mechan Methods 29(1):43–52. https://doi.org/10.1080/15376516.2018.1506851
Duhan M, Singh R, Devi M, Sindhu J, Bhatia R, Kumar A, Kumar P (2019) Synthesis, molecular docking and QSAR study of thiazole clubbed pyrazole hybrid as alpha-amylase inhibitor. J Biomol Struct Dyn:1–17. https://doi.org/10.1080/07391102.2019.1704885
Kumar P, Kumar A (2020) Nucleobase sequence based building up of reliable QSAR models with the index of ideality correlation using Monte Carlo method. J Biomol Struct Dyn 38(11):3296–3306. https://doi.org/10.1080/07391102.2019.1656109
Roy PP, Roy K (2009) QSAR studies of CYP2D6 inhibitor aryloxypropanolamines using 2D and 3D descriptors. Chem Biol Drug Des 73(4):442–455. https://doi.org/10.1111/j.1747-0285.2009.00791.x
Roy K, Mitra I, Kar S, Ojha PK, Das RN, Kabir H (2012) Comparative studies on some metrics for external validation of QSPR models. J Chem Inf Model 52(2):396–408. https://doi.org/10.1021/ci200520g
Golbraikh A, Wang XS, Zhu H, Tropsha A (2012) Predictive QSAR modeling: methods and applications in drug discovery and chemical risk assessment. In: Leszczynski J (ed) Handbook of computational chemistry. Springer Netherlands, Dordrecht, pp 1309–1342. https://doi.org/10.1007/978-94-007-0711-5_37
Gramatica P (2007) Principles of QSAR models validation: internal and external. QSAR Comb Sci 26(5):694–701. https://doi.org/10.1002/qsar.200610151
Nimbhal M, Bagri K, Kumar P, Kumar A (2020) The index of ideality of correlation: a statistical yardstick for better QSAR modeling of glucokinase activators. Struct Chem 31(2):831–839. https://doi.org/10.1007/s11224-019-01468-w
Toropova AP, Toropov AA (2019) Does the index of ideality of correlation detect the better model correctly? Mol Inform 38(8–9):e1800157. https://doi.org/10.1002/minf.201800157
Rucker C, Rucker G, Meringer M (2007) y-Randomization and its variants in QSPR/QSAR. J Chem Inf Model 47 (6):2345–2357. https://doi.org/10.1021/ci700157b
Ojha PK, Mitra I, Das RN, Roy K (2011) Further exploring rm2 metrics for validation of QSPR models. Chemometr Intel Lab Syst 107 (1):194–205. https://doi.org/10.1016/j.chemolab.2011.03.011
Kumar P, Kumar A (2018) Monte Carlo method based QSAR studies of Mer kinase inhibitors in compliance with OECD principles. Drug Res (Stuttg) 68 (4):189–195. https://doi.org/10.1055/s-0043-119288
Dassault-Systèmes-BIOVIA (2019). Discovery Studio, San Diego: Dassault Systèmes. Discovery Studio, San Diego: Dassault Systèmes.,
Acknowledgments
The authors are thankful to Dr. Andrey A. Toropov and Dr. Alla P. Toropova for providing the CORAL software. The authors are also thankful to their respective universities for providing the infrastructure.
Author information
Authors and Affiliations
Contributions
Authors have done equivalent contributions to this work.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 55 kb)
Rights and permissions
About this article
Cite this article
Kumar, A., Kumar, P. Identification of good and bad fragments of tricyclic triazinone analogues as potential PKC-θ inhibitors through SMILES–based QSAR and molecular docking. Struct Chem 32, 149–165 (2021). https://doi.org/10.1007/s11224-020-01629-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11224-020-01629-2