Abstract
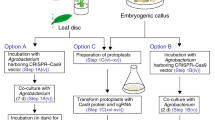
To develop an effective genome editing tool for blueberry breeding, CRISPR-Cas9 and CRISPR-Cas12a were evaluated for their editing efficiencies of a marker gene, beta-glucuronidase (gusA), which was previously introduced into two blueberry cultivars each a single-copy transgene. Four expression vectors were built, with CRISPR-Cas9 and CRISPR-Cas12a each driven by a 35S promoter or AtUbi promoter. Each vector contained two editing sites in the gusA. These four vectors were respectively transformed into the leaf explants of transgenic gusA blueberry and the resulting transgenic calli were induced under hygromycin selection. GUS staining showed that some small proportions of the hygromycin-resistant calli had non-GUS stained sectors, suggesting some possible occurrences of gusA editing. We sequenced GUS amplicons spanning the two editing sites in three blueberry tissues and found about 5.5% amplicons having editing features from the calli transformed with the 35S-Cas9 vector. Further, we conducted a second round of shoot regeneration from leaf explants derived from the initial Cas9- and Cas12a-containing calli (T0) and analyzed amplicons of the target editing region. Of the newly induced shoots, 15.5% for the 35S-Cas9 and 5.3% for the AtUbi-Cas9 showed non-GUS staining, whereas all of the shoots containing the Cas12a vectors showed blue staining. Sanger sequencing confirmed the editing-induced mutations in two representative non-GUS staining lines. Clearly, the second round of regeneration had enriched editing events and enhanced the production of edited shoots. The results and protocol described will be helpful to facilitating high-precision breeding of blueberries using CRISPR Cas technologies.
Key Message
A second round of regeneration enriched editing events and enhanced the production of edited blueberry shoots. The new protocol described facilitates high-precision breeding of blueberries using CRISPR Cas technologies.





Similar content being viewed by others
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Alok A, Sandhya D, Jogam P, Rodrigues V, Bhati KK, Sharma H, Kumar J (2020) The rise of the CRISPR/Cpf1 system for efficient genome editing in plants. Front Plant Sci 11
Ballington JR (2009) The role of interspecific hybridization in blueberry improvement. Acta Hortic 810:49–59
Bandyopadhyay A, Kancharla N, Javalkote VS, Dasgupta S, Brutnell TP (2020) CRISPR-Cas12a (Cpf1): a versatile tool in the plant genome editing tool box for agricultural advancement. Front Plant Sci 11:584151
Bibikova M, Golic M, Golic KG, Carroll D (2002) Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics 161:1169–1175
Bortesi L, Fischer R (2015) The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol Adv 33:41–52
Brevis P, Bassil N, Ballington J, Hancock J (2008) Impact of wide hybridization on highbush blueberry breeding. J Am Soc Hortic Sci 133:11
Charrier A, Vergne E, Dousset N, Richer A, Petiteau A, Chevreau E (2019) Efficient targeted mutagenesis in apple and first time edition of pear using the CRISPR-Cas9 system. Front Plant Sci 10:40
Colle M, Leisner CP, Wai CM, Ou S, Bird KA, Wang J, Wisecaver JH, Yocca AE, Alger EI, Tang H, Xiong Z, Callow P, Ben-Zvi G, Brodt A, Baruch K, Swale T, Shiue L, Song GQ, Childs KL, Schilmiller A, Vorsa N, Buell CR, VanBuren R, Jiang N, Edger PP (2019) Haplotype-phased genome and evolution of phytonutrient pathways of tetraploid blueberry. Gigascience 8:giz012
Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song KM, Pikaard CS (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45:616–629
Ehlenfeldt MK, Prior RL (2001) Oxygen radical absorbance capacity (ORAC) and phenolic and anthocyanin concentrations in fruit and leaf tissues of highbus blueberry. J Agr Food Chem 49:2222–2227
Endo A, Toki S (2019) Targeted mutagenesis using FnCpf1 in Tobacco. Plant Genome Editing with Crispr Systems: Methods and Protocols 1917:269–281
Endo A, Masafumi M, Kaya H, Toki S (2016) Efficient targeted mutagenesis of rice and tobacco genomes using Cpf1 from Francisella novicida. Sci Rep 6:38169
Gallardo RK, Zhang Q, Dossett M, Polashock JJ, Rodriguez-Saona C, Vorsa N, Edger PP, Ashrafi H, Babiker E, Finn CE, Iorizzo M (2018) Breeding trait priorities of the blueberry industry in the United States and Canada. HortScience 53:1021
Gao J, Wang G, Ma S, Xie X, Wu X, Zhang X, Wu Y, Zhao P, Xia Q (2015) CRISPR/Cas9-mediated targeted mutagenesis in Nicotiana tabacum. Plant Mol Biol 87:99–110
Gao W, Long L, Tian X, Xu F, Liu J, Singh PK, Botella JR, Song C (2017) Genome editing in cotton with the CRISPR/Cas9 system. Front Plant Sci 8:1364
Goyali JC, Igamberdiev AU, Debnath SC (2018) DNA methylation in lowbush blueberry (Vaccinium angustifolium Ait.) propagated by softwood cutting and tissue culture. Can J Plant Sci 98:1035–1044
Gupta V, Estrada AD, Blakley IC, Reid R, Patel K, Meyer MD, Andersen SU, Brown AF, Lila MA, Loraine A (2015) RNASeq analysis and annotation of a draft blueberry genome assembly identifies candidate genes involved in fruit ripening, biosynthesis of bioactive compounds, and stage-specific alternative splicing. GigaScience 4:5
Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, MacManes MD, Ott M, Orvis J, Pochet N, Strozzi F, Weeks N, Westerman R, William T, Dewey CN, Henschel R, Leduc RD, Friedman N, Regev A (2013) De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc 8:1494–1512
Hood EE, Gelvin SB, Melchers LS, Hoekema A (1993) NewAgrobacterium helper plasmids for gene transfer to plants. Transgenic Res 2:208–218
Hooghvorst I, Lopez-Cristoffanini C, Nogues S (2019) Efficient knockout of phytoene desaturase gene using CRISPR/Cas9 in melon. Sci Rep 9
Jefferson RA, Kavanagh TA, Bevan MW (1987) Gus fusions - beta-glucuronidase as a sensitive and versatile gene fusion marker in higher-plants. Embo J 6:3901–3907
Jeggo PA (1998) DNA breakage and repair. Adv Genet 38:185–218
Jia H, Orbovic V, Wang N (2019) CRISPR-LbCas12a-mediated modification of citrus. Plant Biotechnol J 17:1928–1937
Khanna KK, Jackson SP (2001) DNA double-strand breaks: signaling, repair and the cancer connection. Nat Genet 27:247–254
Kim H, Kim ST, Ryu J, Kang BC, Kim JS, Kim SG (2017) CRISPR/Cpf1-mediated DNA-free plant genome editing. Nat Commun 8
LeBlanc C, Zhang F, Mendez J, Lozano Y, Chatpar K, Irish VF, Jacob Y (2018) Increased efficiency of targeted mutagenesis by CRISPR/Cas9 in plants using heat stress. Plant J 93:377–386
Lee K, Zhang Y, Kleinstiver BP, Guo JA, Aryee MJ, Miller J, Malzahn A, Zarecor S, Lawrence-Dill CJ, Joung JK, Qi Y, Wang K (2019) Activities and specificities of CRISPR/Cas9 and Cas12a nucleases for targeted mutagenesis in maize. Plant Biotechnol J 17:362–372
Li B, Rui H, Li Y, Wang Q, Alariqi M, Qin L, Sun L, Ding X, Wang F, Zou J, Wang Y, Yuan D, Zhang X, Jin S (2019) Robust CRISPR/Cpf1 (Cas12a)-mediated genome editing in allotetraploid cotton (Gossypium hirsutum). Plant Biotechnol J 17:1862–1864
Lin TY, Walworth A, Zong XJ, Danial GH, Tomaszewski EM, Callow P, Han XM, Zaharia LI, Edger PP, Zhong GY, Song GQ (2019) VcRR2 regulates chilling-mediated flowering through expression of hormone genes in a transgenic blueberry mutant. Hortic Res-England 6
Liu HY, Wang K, Jia ZM, Gong Q, Lin ZS, Du LP, Pei XW, Ye XG (2020) Efficient induction of haploid plants in wheat by editing of TaMTL using an optimized Agrobacterium-mediated CRISPR system. J Exp Bot 71:1337–1349
Lowder LG, Zhang D, Baltes NJ, Paul JW 3rd, Tang X, Zheng X, Voytas DF, Hsieh TF, Zhang Y, Qi Y (2015) A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol 169:971–985
Mahfouz MM (2017) Genome editing: the efficient tool CRISPR-Cpf1. Nat Plants 3:17028
Malzahn AA, Tang X, Lee K, Ren Q, Sretenovic S, Zhang Y, Chen H, Kang M, Bao Y, Zheng X, Deng K, Zhang T, Salcedo V, Wang K, Zhang Y, Qi Y (2019) Application of CRISPR-Cas12a temperature sensitivity for improved genome editing in rice, maize, and Arabidopsis. BMC Biol 17:9
Mao YF, Botella JR, Liu YG, Zhu JK (2019) Gene editing in plants: progress and challenges. Natl Sci Rev 6:421–437
Nakajima I, Ban Y, Azuma A, Onoue N, Moriguchi T, Yamamoto T, Toki S, Endo M (2017) CRISPR/Cas9-mediated targeted mutagenesis in grape. PLoS ONE 12:e0177966
Neto CC (2007) Cranberry and blueberry: evidence for protective effects against cancer and vascular diseases. Mol Nutr Food Res 51:652–664
Norris SR, Barrette TR, DellaPenna D (1995) Genetic dissection of carotenoid synthesis in arabidopsis defines plastoquinone as an essential component of phytoene desaturation. Plant Cell 7:11
Omori M, Yamane H, Osakabe K, Osakabe Y, Tao R (2021) Targeted mutagenesis of CENTRORADIALIS using CRISPR/Cas9 system through the improvement of genetic transformation efficiency of tetraploid highbush blueberry. J Hortic Sci Biotechnol 96:153–161
Osakabe Y, Liang Z, Ren C, Nishitani C, Osakabe K, Wada M, Komori S, Malnoy M, Velasco R, Poli M, Jung MH, Koo OJ, Viola R, Nagamangala Kanchiswamy C (2018) CRISPR-Cas9-mediated genome editing in apple and grapevine. Nat Protoc 13:2844–2863
Park J, Lim K, Kim JS, Bae S (2017) Cas-analyzer: an online tool for assessing genome editing results using NGS data. Bioinformatics 33:286–288
Qin G, Gu H, Ma L, Peng Y, Deng XW, Chen Z, Qu LJ (2007) Disruption of phytoene desaturase gene results in albino and dwarf phenotypes in Arabidopsis by impairing chlorophyll, carotenoid, and gibberellin biosynthesis. Cell Res 17:471–482
Rowland LJ, Nguyen B (1993) Use of polyethylene-glycol for purification of DNA from leaf tissue of woody-plants. Biotechniques 14:734–740
Schindele P, Puchta H (2020) Engineering CRISPR/LbCas12a for highly efficient, temperature-tolerant plant gene editing. Plant Biotechnol J 18:1118–1120
Shan Q, Wang Y, Li J, Gao C (2014) Genome editing in rice and wheat using the CRISPR/Cas system. Nat Protoc 9:2395–2410
Song GQ (2015) Blueberry (Vaccinium corymbosum L.). Methods Mol Biol 1224:121–131
Song G-Q, Chen Q (2018) Overexpression of the MADS-box gene K-domain increases the yield potential of blueberry. Plant Sci 276:10
Song G, Sink K (2004) Agrobacterium tumefaciens-mediated transformation of blueberry (Vaccinium corymbosum L.). Plant Cell Rep 23:475–484
Song GQ, Prieto H, Orbovic V (2019a) Agrobacterium-mediated transformation of tree fruit crops: methods, progress, and challenges. Front Plant Sci 10:226
Song GQ, Walworth A, Lin T, Chen Q, Han X, Irina Zaharia L, Zhong GY (2019b) VcFT-induced mobile florigenic signals in transgenic and transgrafted blueberries. Hortic Res 6:105
Surridge C (2019) Blueberry fooled into flowering. Nat Plants 5:910–910
Tang X, Lowder LG, Zhang T, Malzahn AA, Zheng X, Voytas DF, Zhong Z, Chen Y, Ren Q, Li Q, Kirkland ER, Zhang Y, Qi Y (2017) A CRISPR-Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat Plants 3:17018
Vander Kloet SP (1988) The genus Vaccinium in North America. Res Branch Agric Can Publ 1828:201
Vu TV, Sivankalyani V, Kim EJ, Doan DTH, Tran MT, Kim J, Sung YW, Park M, Kang YJ, Kim JY (2020) Highly efficient homology-directed repair using CRISPR/Cpf1-geminiviral replicon in tomato. Plant Biotechnol J 18:2133–2143
Walworth A, Song GQ (2018) The cold-regulated genes of blueberry and their response to overexpression of VcDDF1 in several tissues. Int J Mol Sci 19
Wang Z, Wang S, Li D, Zhang Q, Li L, Zhong C, Liu Y, Huang H (2018) Optimized paired-sgRNA/Cas9 cloning and expression cassette triggers high-efficiency multiplex genome editing in kiwifruit. Plant Biotechnol J 16:1424–1433
Xu R, Qin R, Li H, Li D, Li L, Wei P, Yang J (2017) Generation of targeted mutant rice using a CRISPR-Cpf1 system. Plant Biotechnol J 15:713–717
Yin X, Biswal AK, Dionora J, Perdigon KM, Balahadia CP, Mazumdar S, Chater C, Lin HC, Coe RA, Kretzschmar T, Gray JE, Quick PW, Bandyopadhyay A (2017) CRISPR-Cas9 and CRISPR-Cpf1 mediated targeting of a stomatal developmental gene EPFL9 in rice. Plant Cell Rep 36:745–757
Zaidi SS, Mahfouz MM, Mansoor S (2017) CRISPR-Cpf1: a new tool for plant genome editing. Trends Plant Sci 22:550–553
Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, Koonin EV, Zhang F (2015) Cpf1 Is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163:759–771
Zetsche B, Heidenreich M, Mohanraju P, Fedorova I, Kneppers J, DeGennaro EM, Winblad N, Choudhury SR, Abudayyeh OO, Gootenberg JS, Wu WY, Scott DA, Severinov K, van der Oost J, Zhang F (2017) Multiplex gene editing by CRISPR-Cpf1 using a single crRNA array. Nat Biotechnol 35:31–34
Zhong ZH, Zhang YX, You Q, Tang X, Ren QR, Liu SS, Yang LJ, Wang Y, Liu XP, Liu BL, Zhang T, Zheng XL, Le Y, Zhang Y, Qi YP (2018) Plant genome editing using FnCpf1 and LbCpf1 nucleases at redefined and altered PAM sites. Mol Plant 11:999–1002
Funding
This research was supported in part by the agreement of Non-Assistance Cooperative Agreement #58-8060-6-009 between the USDA Agricultural Research Service and Michigan State University. Dr. Xiaoyan Han’s visit at Michigan State University was partially supported by a CAS Scholarship. Dr. Emadeldin A. H. Ahmed is a Fulbright fellow at Michigan State University.
Author information
Authors and Affiliations
Contributions
GS and GZ conceived and supervised the research. YY, XYH, XH, JR, EA, and GS conducted the experiments. YY, XYH, and YQ analyzed the data. GS, XYH, YY, YQ, and GZ wrote the manuscript. All authors reviewed and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that no competing interests exist.
Additional information
Communicated by Joyce Van Eck.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.



Rights and permissions
About this article
Cite this article
Han, X., Yang, Y., Han, X. et al. CRISPR Cas9- and Cas12a-mediated gusA editing in transgenic blueberry. Plant Cell Tiss Organ Cult 148, 217–229 (2022). https://doi.org/10.1007/s11240-021-02177-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-021-02177-1