Abstract
Mediterranean diet has been proposed to promote healthy aging, but its effects on aging biomarkers have been poorly investigated. We evaluated the impact of a 1-year Mediterranean-like diet in a pilot study including 120 elderly healthy subjects from the NU-AGE study (60 Italians, 60 Poles) by measuring the changes in their epigenetic age, assessed by Horvath’s clock. We observed a trend towards epigenetic rejuvenation of participants after nutritional intervention. The effect was statistically significant in the group of Polish females and in subjects who were epigenetically older at baseline. A genome-wide association study of epigenetic age changes after the intervention did not return significant (adjusted p value < 0.05) loci. However, we identified small-effect alleles (nominal p value < 10–4), mapping in genes enriched in pathways related to energy metabolism, regulation of cell cycle, and of immune functions. Together, these findings suggest that Mediterranean diet can promote epigenetic rejuvenation but with country-, sex-, and individual-specific effects, thus highlighting the need for a personalized approach to nutritional interventions.
Similar content being viewed by others
Introduction
As the population continues to age within Europe, an increase in the incidence of age-related diseases is observed (WHO 2017). Indeed, the increasing lifespan is not associated with an increase in health span, and this issue represents a great challenge for our societies. There is an important need to identify factors that are able to influence health in old age and to develop and validate interventions that could slow down or counteract the process of aging and its associated pathologies. A possible strategy to impact on aging is to intervene on lifestyle factors, such as diet or physical activity. Nutritional interventions seem to be one of the most promising approaches to promote healthy aging, and growing amount of data indicates that they can influence the health status of subjects (Longo et al. 2015; Dato et al. 2016; Wahl et al. 2016; Shlisky et al. 2017; Xia et al. 2017; Heiss et al. 2017).
Mediterranean diet, which is considered by UNESCO as a heritage of humanity, is a well-balanced mix of nutrients, anti-oxidants, and anti-inflammatory molecules, and it has been recently suggested that some of its components are able to exert hormetic effects (Martucci et al. 2017). This diet has demonstrated favorable effects on cardiovascular risk, blood pressure, cancer, inflammation, or frailty status (Estruch et al. 2006, 2013; Mitjavila et al. 2013; Ostan et al. 2015; Martínez-González et al. 2015; Kojima et al. 2018), and it has been observed that it can impact methylation of inflammation-related genes in peripheral blood cells (Arpón et al. 2017). Some studies have suggested that Mediterranean diet prevents telomere shortening, a well-established biomarker of age, but results are not consistent among different studies (Davinelli et al. 2019). The role of Mediterranean diet in promoting healthy aging has been recently investigated in the framework of the European project NU-AGE (“New dietary strategies addressing the specific needs of elderly population for an healthy aging in Europe” (http://www.nu-age.eu/)), a large multidisciplinary consortium with 30 partners across Europe (Berendsen et al. 2014; Santoro et al. 2014). The aim of NU-AGE project was to investigate how an intervention based on Mediterranean diet, specifically tailored according to the nutritional needs of people over 65 years of age, can impact on age-related diseases and functional decline, possibly modulating inflammaging and its outcomes (Franceschi et al. 2000). Probands were enrolled in five European countries (Italy, Poland, France, the Netherlands, and the UK), and a 1-year Mediterranean-like diet was administered to the intervention subgroup. A comprehensive clinical and molecular characterization was performed at baseline and after the 1-year intervention, and results achieved so far in the framework of this study have demonstrated a beneficial effect of the Mediterranean-like diet on global cognition and episodic memory (Marseglia et al. 2018), osteoporosis (Jennings et al. 2018), immune function (Maijo et al. 2018), and cardiovascular health (Jennings et al. 2019), as well as on the proteasomal proteolysis (Athanasopoulou et al. 2018). The NU-AGE study design (different countries with different dietary traditions and habits) and the large number of collected data allowed to evaluate the impact of relevant variables usually poorly investigated (age, sex, and ethnicity/genetics, as well as individual characteristics) on different parameters at baseline and after the intervention (Konz et al. 2018; Marseglia et al. 2018; Ostan et al. 2018; Pujos-Guillot et al. 2018; Santoro et al. 2018, 2019; Jennings et al. 2019). Importantly, the enrolled subjects were recruited also in non-Mediterranean countries (Poland, the Netherlands, and the UK). The effects of a Mediterranean diet intervention on non-Mediterranean countries is not granted, because its transferability requires specific changes in dietary habits (Martínez-González et al. 2017) and because genetic and environmental factors, that can be country-specific, can hamper/enhance its effects (Mayr et al. 2018).
In order to monitor the impact of anti-aging interventions, accurate biological measures of age are needed. The discovery of the so-called epigenetic clocks, based on DNA methylation (DNAm) levels at some specific sites, has been a major breakthrough in this field within the last 6 years (Horvath 2013; Hannum et al. 2013; Weidner et al. 2014). These biomarkers have been proposed as accurate and robust biomarkers of aging, but also as indicators of the biological health of an individual: DNAm age, also known as epigenetic age, measured in blood cells, has been found to be predictive of mortality (Marioni et al. 2015a; Perna et al. 2016; Christiansen et al. 2016; Chen et al. 2016; Dugué et al. 2018) and other aging-related outcomes such as frailty (Breitling et al. 2016) or cognitive and physical functioning (Marioni et al. 2015b; Degerman et al. 2017; Simpkin et al. 2017; Gale et al. 2018a; Sillanpää et al. 2018). Recent studies demonstrated that age-associated epigenetic variations can be affected by diet (Bacalini et al. 2014; Kok et al. 2015; Quach et al. 2017) and that exposure to some pathogenic conditions or environmental factors can influence DNAm age (Horvath et al. 2014; Nevalainen et al. 2017; Li et al. 2018; Rosen et al. 2018); however, little is known about the specific relationship between nutritional interventions and epigenetic biomarkers of aging.
In the present manuscript, we used epigenetic biomarkers of aging to study the impact of the nutritional intervention delivered in the framework of NU-AGE project, focusing on subjects enrolled in a Mediterranean country (Italy) and in a non-Mediterranean country (Poland).
Subjects and methods
NU-AGE study
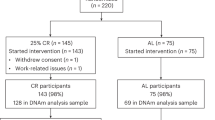
NU-AGE was a 1-year, multicenter, randomized, single-blind, controlled trial (registered with clinicaltrials.gov, NCT01754012) with two parallel groups (i.e., dietary intervention and control) carried out during April 2012–January 2015 in five European centers in Italy, Poland, France, the Netherlands, and the UK. The recruitment of participants has been described in detail previously (Berendsen et al. 2014; Santoro et al. 2014). Briefly, 1279 participants aged 65–79 years, free of major overt chronic diseases for at least 2 years (i.e., cancer, severe organ disease), living independently, and free of dementia, were recruited to participate in the baseline assessment. At enrollment, exclusion criteria included severe heart diseases, type 1 and insulin-treated type 2 diabetes, chronic use of corticosteroids, recent use of antibiotics, change in habitual medication use, frailty (Fried et al. 2001), malnutrition (body mass index (BMI) < 18.5 kg/m2 or 10% weight loss within 6 months), or food allergy/intolerance requiring special diets. Participants were randomly assigned (1:1) to the control or intervention groups, after stratification by sex, age (65–72 or > 72–79 years), frailty status (pre-frail or non-frail), and body mass index (< 25 or ≥ 25 kg/m2). All participants provided written informed consent.
Epigenetic analysis was performed in a subgroup of 120 randomly selected subjects (60 from the Italian cohort and 60 from the Polish one) from the intervention group, at both baseline (T0) and after 1 year of dietary intervention (T1), for a total of 240 samples. Exact chronological age of the subjects (in years) at T0 was calculated as follows: [(T0 date) − (date of birth)]/365. Exact chronological age at T1 was calculated as follows: [(T1 date) − (date of birth)]/365. Adherence to study protocol was evaluated using the 7-day food records. A NU-AGE index scoring system was specifically developed and used as a measure of adherence to the NU-AGE diet, as described previously (Jennings et al. 2018).
Analysis of DNA methylation
Samples were analyzed for genome-wide DNA methylation patterns using the Illumina Infinium HumanMethylation450 BeadChip array (Illumina Inc., CA, USA). Genomic DNA was extracted from 250 μL of whole blood (drawn on EDTA tubes), using the QIAamp 96 DNA Blood Kit (QIAGEN, Hilden, Germany). One microgram of DNA was bisulfite converted, using the EZ DNA Methylation Kit (Zymo Research Corporation, Orange, CA, USA) according to manufacturer’s instructions. After bisulfite conversion, DNA was whole-genome amplified, enzymatically fragmented, and hybridized to the Illumina Infinium HumanMethylation450 BeadChips (Illumina Inc., CA, USA), according to the manufacturer’s protocols. Samples from the different groups (Italy and Poland, T0 and T1) were accurately randomized across the experimental sessions. Arrays were scanned using the HiScan instrument (Illumina Inc., CA, USA). Raw fluorescence intensities were extracted using minfi Bioconductor package, and normalization was performed using the preprocessQuantile function (Touleimat and Tost 2012).
Evaluation of DNA methylation age and of epigenetic age acceleration
Normalized DNA methylation data were uploaded into the DNA methylation age calculator, freely available at the website: https://dnamage.genetics.ucla.edu, to calculate DNAm age, as described by Horvath (Horvath 2013). DNAm age is calculated using the weighted average of DNA methylation levels at 353 CpG sites (Horvath 2013). The “advanced blood analysis” option was selected in the online calculator, allowing the calculation of three measures of epigenetic age acceleration (AA) that were further considered in this study. These measures have been previously described by Horvath and colleagues and have been applied to date in several publications (Levine et al. 2015; Horvath and Ritz 2015; Horvath et al. 2015, 2016; Chen et al. 2016; Ambatipudi et al. 2017; Quach et al. 2017; Maierhofer et al. 2017; Gale et al. 2018a, 2018b). The first measure is considered as the universal measure of epigenetic AA and is denoted AgeAccel. It corresponds to the residual that results from regressing DNAm age on chronological age. The second measure of epigenetic AA is referred as Intrinsic Epigenetic Age Acceleration (IEAA), denoted as AAHOAdjCellCounts in the online software. IEAA is defined as the residual resulting from regressing DNAm age on chronological age and seven measures of immune blood cell count estimates: naive CD8+ T cells, exhausted CD8+ T cells, plasma B cells, CD4+ T cells, natural killer cells, monocytes, and granulocytes. IEAA is independent of changes in blood cell composition that occur with time and is considered as a measure of “pure” epigenetic aging effects in blood cells. Finally, the third measure considered is referred as Extrinsic Epigenetic Age Acceleration (EEAA), known as BioAge4HAStaticAdjAge in the online software. EEAA is based on a weighted average of the epigenetic age measure with Hannum’s clock (Hannum et al. 2013) and three blood cell types that are known to change with age: naive cytotoxic T lymphocytes (CD45RA+CCR7+), exhausted cytotoxic T lymphocytes (CD45RA-CD28-), and plasma B cells. EEAA is defined as the residual formed from regressing the resulting weighted epigenetic age on chronological age. This measure is dependent on age-related changes in blood cell composition and can be considered as a measure of aging in immune system.
Estimating blood cell counts based on DNA methylation levels
Blood cell counts used in the measures of IEAA and EEAA were estimated based on DNA methylation data using the epigenetic clock online software. Blood cell proportions of CD8+ T cells, CD4+ T cells, natural killer cells, B cells, and granulocytes are based on Houseman’s estimation method (Houseman et al. 2012). An advanced analysis option of the epigenetic clock software is used to estimate the percentage of naïve and exhausted CD8+ T cells.
Genotyping
Genomic DNA was extracted from 250 μL of whole blood (drawn on EDTA tubes), using the QIAamp 96 DNA Blood Kit (QIAGEN, Hilden, Germany). Two hundred nanograms of genomic DNA were genotyped for 713,014 genetic markers by the Illumina OmniExpress BeadChip (Illumina Inc., CA, USA), according to manufacturer’s protocol. After quality control, 118 samples were retained. Quantitative trait association analysis and estimation of single nucleotide polymorphisms’ (SNPs) allele frequencies were performed using PLINK toolset.
Statistical analysis
The effects of the nutritional intervention on the three abovementioned measures of epigenetic AA (AgeAccel, IEAA, and EEAA) were analyzed with a Student’s paired-sample t test. For each epigenetic AA measure, Benjamini-Hochberg procedure was applied to correct for multiple tests, considering a total of 6 tests. Pearson correlations between measures of epigenetic age and chronological age or scores of adherence to Mediterranean diet were calculated. All statistical analyses and graphics were produced using the R v3.3.2.
Results
Subjects
Genome-wide DNA methylation profiles were analyzed by the Illumina Infinium HumanMethylation450 Beadchip (Illumina Inc., CA, USA) in whole blood of 120 European subjects belonging to the intervention group of the NU-AGE study, with chronological age ranging from 65 to 79 years old. Sixty patients were recruited in Italy and the other half was recruited in Poland. Characteristics of enrolled subjects are summarized in Table 1. Baseline characteristics were similar between the two groups in terms of chronological age and adherence to Mediterranean diet (Student’s t test p value > 0.05) (Supplemental Fig. 1). Body mass index (BMI) tended to be higher in Polish subjects compared with Italian ones, and this difference was statistically significant when considering only males (Student’s t test p value = 0.027) (Supplemental Fig. 1). After 1 year of nutritional intervention (T1), adherence to Mediterranean diet significantly increased in both Italian and Polish participants, and a significant decrease in BMI was observed in Italian males (paired Student’s t test p value = 0.008) (Supplemental Figs. 1 and 2).
Effect of the nutritional intervention on the epigenetic age acceleration measures
Epigenetic age (also referred to as DNA methylation age (DNAm age)) was calculated using the online age calculator freely available at the website: https://dnamage.genetics.ucla.edu. As expected, DNAm age was significantly associated with chronological age (p < 0.0001), both at T0 (before nutritional intervention) and at T1 (after a 12-month Mediterranean-like nutritional intervention), in both Italian and Polish groups (Fig. 1).
For each subject, we evaluated the epigenetic age acceleration (AA), that is the deviation between DNAm age and effective chronological age. Positive values of epigenetic AA indicate an epigenetic age older than expected, while negative values indicate an epigenetic age younger than expected on the basis of chronological age. In particular, we considered three measures of epigenetic AA, implemented in the online age calculator, which reflect different aspects of epigenetic aging (see the “Subjects and methods” section): (1) AgeAccel; (2) Intrinsic Epigenetic Age Acceleration (IEAA); (3) Extrinsic Epigenetic Age Acceleration (EEAA) (Supplemental Table 1). At T0, in Italian subjects, AgeAccel ranged from − 12.38 to 15.62 years, IEAA ranged from − 11.56 to 12.27 years, and EEAA ranged from − 10.90 to 7.41 years. In Polish subjects, AgeAccel ranged from − 8.49 to 16.27 years, IEAA ranged from − 9.01 to 15.55 years, and EEAA ranged from − 12.56 to 13.56 years. At T1, in Italian subjects, AgeAccel ranged from − 8.60 to 14.25 years, IEAA ranged from − 8.62 to 9.92 years, and EEAA ranged from − 8.97 to 7.90 years. In Polish subjects, at T1, AgeAccel ranged from − 9.81 to 10.99 years, IEAA ranged from − 10.44 to 9.47 years, and EEAA ranged from − 11.31 to 10.02 years. Baseline measures of epigenetic AA were similar between Italian and Polish subjects (Student’s t test p value > 0.05), but EEAA was significantly higher in Polish males compared with Polish females (Student’s t test p = 0.00009) and compared with Italian males (Student’s t test p = 0.02).
In both Italian and Polish cohorts, epigenetic AA measures at baseline were significantly associated (p < 0.05) with those obtained after the 12-month-tailored nutritional intervention (Supplemental Fig. 3).
We then used Student’s paired-sample t test to compare the epigenetic AA measures at T0 and at T1. In Italian subjects, no statistically significant differences between T0 and T1 were observed considering AgeAccel, also when subjects were divided according to sex (Fig. 2, upper panel). On the contrary, in Polish subjects, AgeAccel measures were significantly lower at T1 versus baseline (T0) (p = 0.0312) (Fig. 2, upper panel). In other words, under the Mediterranean-like diet intervention, there was a statistically significant rejuvenation of the Polish subjects, according to the AgeAccel measure. When we divided samples on the basis of sex, we observed that the effect was predominantly related to a decrease in AgeAccel measures in Polish females at T1 compared with T0 (p = 0.0013). Rejuvenation of the Polish females after 1 year of nutritional intervention was confirmed with the IEAA measure (Fig. 2, middle panel), as analysis returned a significant decrease in IEAA values at T1 versus T0 (p = 0.007). Lower IEAA measures were also observed at T1 in Italian subjects as compared with T0 (p = 0.0347). The EEAA predictor did not give significant results (Fig. 2, lower panel) in both groups. After correction for multiple testing, the effect remained statistically significant for Polish females, according to AgeAccel (adjusted p value = 0.008) and IEAA (adjusted p value = 0.04) measures (Fig. 2).
A 1 year Mediterranean-like diet intervention promotes epigenetic rejuvenation in a country- and sex-specific manner. Boxplots of epigenetic AA measures at T0 and T1 (upper panel, AgeAccel; middle panel, IEAA; lower panel, EEAA) considering all the subjects, only males, and only females. Uncorrected p values and p values after correction for multiple testing are displayed.
Supplemental Fig. 4 reports, for each subject, the intra-pair difference between AgeAccel at T1 and AgeAccel at T0 (AgeAccel Diff), the intra-pair difference between IEAA at T1 and IEAA at T0 (IEAA Diff), and the intra-pair difference between EEAA at T1 and EEAA at T0 (IEAA Diff). In all three cases, a negative value indicates an epigenetic rejuvenation.
Finally, we assessed if AgeAccel Diff, IEAA Diff, and EEAA Diff values were related to, respectively, Age Accel, IEAA, and EEAA values at baseline (Fig. 3). In both the countries and for all the different AA measures, we found that the majority of subjects showing an epigenetic age rejuvenation (AgeAccel Diff, IEAA Diff, or EEAA Diff less than 0) had also baseline AA levels greater than 0 (Fig. 3). Fisher’s exact test confirmed that this enrichment was significant for AgeAccel and IEAA measures in Poles, indicating that the effect of the diet tended to be more marked in those subjects that displayed higher epigenetic AA values at T0.
Stronger impact of diet on epigenetic AA measures in subjects with higher epigenetic AA values at baseline. Scatter plots of epigenetic AA measures at T1 (x-axis) versus epigenetic AA measures difference between T1-T0 (y-axis). The three epigenetic AA measures (AgeAccel, IEAA, and EEEA) are reported respectively in the upper, middle, and lower panels. Males and females subjects are indicated with cyan and pink circles, respectively. The number of subjects in each quadrant is reported. Fisher’s exact test was applied to test if there was a difference of proportion of subjects in the 4 quadrants
Association between epigenetic age acceleration measures, BMI, and adherence to the Mediterranean-like diet
In order to identify factors associated with the slowdown of the epigenetic AA measures, we first investigated the relationship between BMI and the epigenetic markers. We did not find any significant association between BMI and AgeAccel, IEEA or EEAA (results not shown).
We also analyzed the association between the epigenetic AA measures and the NU-AGE score measuring the adherence to the Mediterranean-like diet, calculated at T0 and T1 (see the “Subjects and methods” section). We observed a significant negative association of AgeAccel (p = 0.037) and IEAA (p = 0.027) with the NU-AGE score, with higher levels of adherence to the Mediterranean-like diet associated with negative epigenetic AA values, that is with epigenetic rejuvenation (Supplemental Fig. 5).
Association between epigenetic age acceleration measures and genotype
Finally, we evaluated if a response to Mediterranean-like dietary intervention, in terms of epigenetic AA, was related to the genetic background of the participants of the study. To this aim, we carried out a genome-wide association study (GWAS) of epigenetic AA measures in our cohort, expressed as AgeAccel Diff, IEAA Diff, or EEAA Diff as described above (Supplemental Fig. 4). The quantile-quantile (QQ) plot of association results demonstrated no genomic inflation (data not shown). After correction for multiple testing, no significant association was observed at the genome-wide level (Benjamini-Hochberg corrected p value < 0.05). However, small-effect loci with nominal significance (p value < 1 × 10−4) were identified for all of the three measures of epigenetic AA. A total of 68, 49, and 46 single nucleotide polymorphisms (SNPs) were found significantly associated with AgeAccel Diff, IEAA Diff, and EEAA Diff, respectively (Supplemental Table 2). Thirty-one SNPs were common between AgeAccel Diff and IEAA Diff, while there were no SNPs in common between EEAA Diff and AgeAccel Diff or IEAA Diff (Fig. 4). Interestingly, 5 SNPs out of 68 (for AgeAccel Diff) and 6 SNPs out of 49 (for IEAA Diff) showed minor allele frequency differences between Italians and Polish (p values < 0.05).
In order to identify pathways that may be relevant to epigenetic AA effects upon Mediterranean-like nutritional intervention, we performed enrichment analysis using i-GSEA4GWAS (Supplemental Table 3). In the analysis of AgeAccel Diff associations, we found 60 significant gene sets (p < 0.05); 13 of which were significant after false discovery rate (FDR) correction (FDR < 0.05). IEAA Diff analysis returned 37 significant gene sets (p < 0.05); 11 of which had a FDR < 0.05. We found a large overlap between the enrichment analysis results of the two epigenetic AA measures, in particular for pathways involved in energy metabolism, regulation of cell cycle, and of immune functions. On the contrary, enrichment analysis for EEAA Diff did not return any significant result (p < 0.05).
Discussion
In this study, we observed the effects of a 1-year Mediterranean-like diet, newly designed according to the nutritional needs of people over 65 years of age (Berendsen et al. 2014), on epigenetic AA measures. We analyzed blood methylation data, obtained in a population of subjects enrolled in a multicenter trial, and we demonstrated that the nutritional intervention delivered in the NU-AGE study can slow down the epigenetic aging rate of blood in specific groups of participants.
It is known that environmental factors, including diet, are able to modify the epigenome (Bacalini et al. 2014), and cross-sectional associations between epigenetic AA measures and diet have been previously described by Quach et al. (Quach et al. 2017). However, data from longitudinal studies on a possible rejuvenation of epigenetic age with dedicated therapeutic or lifestyle interventions are few. Only two works have been recently published on this topic. Firstly, Pavanello et al. examined the hypothesis that an intensive relaxing training of 60 days may influence epigenetic age by turning back the epigenetic clock (Pavanello et al. 2019) (Pavanello et al. 2019). They observed a trend to a reduction in DNAm age (estimated with the model proposed by Zbiec-Piekarska et al. (Zbieć-Piekarska et al. 2015) after training in six healthy subjects, but the effect was not statistically significant (p = 0.053). Secondly, the effect of a protocol intended to “rejuvenate the thymus” (thymus regeneration, immunorestoration, and insulin mitigation, TRIIM trial) was examined by Horvath’s team in a small, non-controlled study (Fahy et al. 2019). The 1-year intervention, composed of recombinant human growth hormone, dehydroepiandrosterone, and metformin, was delivered to 9 healthy aging men (age range 51–65 years old). A rejuvenating effect on four epigenetic age predictors (Horvath 2013; Hannum et al. 2013; Levine et al. 2018; Lu et al. 2019) was observed, with a mean change of about 2.5 years. The intervention was also associated with a protective effect on different immunosenescence biomarkers (reversal of thymic involution, increase in both naïve CD4+ and CD8+ T cells), and the effect persisted 6 months after discontinuing the treatment (Fahy et al. 2019).
Data regarding the impact of nutritional intervention are lacking, and to our knowledge, our study is the first longitudinal and interventional study to examine effects of such an intervention on epigenetic age acceleration measures in human blood cells. According to our results, a 1-year nutritional intervention could be able to rewind the epigenetic AA process in some specific groups. The discrepancy between the slowdown obtained with AgeAccel and IEAA measures on one hand, and the absence of effect observed with EEAA measure on the other hand seems to be of particular interest. Indeed, the three measures of epigenetic AA we studied in this work do not capture the same features of biological aging. By their very own construction, IEAA is considered as a measure of epigenetic age acceleration independent of age-related changes in the cellular composition of blood, whereas EEAA is more meant to capture the age-related decline of the immune system. Here, we did not observe any significant impact of the nutritional intervention on this decline according to the EEAA measure. Our results therefore suggest that the Mediterranean-like diet has a pure rejuvenating impact on the biological clock, and that this result is unconfounded by a potential effect of the intervention on the immune system.
Interestingly, the protective effect of the whole diet on the epigenetic age appears to be both country- and sex-specific, as Polish, and especially Polish females, appear to benefit the more for the intervention, according to the measures of epigenetic AA. Epigenetic aging rates have been previously described as influenced by race/ethnicity (Horvath et al. 2016) and sex (Horvath et al. 2016; Xiao et al. 2018), and we also demonstrated here that the epigenetic response to an intervention can be influenced by these parameters. It is likely that the observed differences between males and females are related not only to pure biological differences (for example, differences in body composition (Santoro et al. 2018), but also to anthropologic and cultural components (such as levels of education, cooking, or willingness to stick to the nutritional advices for example).
While population and sex-specificities appear clearly in this work, inter-individual differences intervene also in the response to the nutritional intervention. Firstly, subjects that were epigenetically older at baseline (i.e., subjects with higher epigenetic AA values at T0) had a more marked effect of the nutritional intervention and seemed to benefit more of the effects of the Mediterranean-like diet. Secondly, according to our GWAS, some genetic variants influence the response to the intervention. GWAS results were largely overlapping between AgeAccel and IEAA analysis. Furthermore, enrichment analysis suggested that both epigenetic AA measures were associated to genetic variants in genes involved in pathways related to the regulation of cell metabolism and immune function. Among the gene sets associated to AgeAccel differences between T1 and T0, it is worth to note the presence of the mTOR pathway, which plays a pivotal role in the regulation of nutrients-sensing and energy metabolism during aging (Cummings and Lamming 2017; Tosti et al. 2018; Lushchak et al. 2019). In animal models, it has been previously demonstrated the influence of a genetic component in the response to a nutritional intervention, such as caloric restriction (CR) (Liao et al. 2010). Liao et al. observed that lifespan expansion by CR was not universal in mice and was highly dependent on the strain of the animals, suggesting the important influence of the genotype in the CR effect (Liao et al. 2010). In humans, this problem has been poorly investigated. Previous reports have evaluated the association between genetic background and epigenetic AA in different tissues (Lu et al. 2016, 2017, 2018), but little is known about the influence of genetics on the response to a nutritional intervention. In a recent study, the effects of dietary supplementation with folic acid and vitamin B12 on epigenetic age deceleration were found dependent upon gender and MTHFR genotype (Sae-Lee et al. 2018). Only the group of women with the MTHFR 677CC genotype displayed a deceleration in epigenetic aging upon vitaminic supplementation (Sae-Lee et al. 2018).
In summary, strengths of our study include the following: (1) the experimental design (multi-center trial with 1-year follow-up), with the inclusion of two different populations coming from one Mediterranean and one non-Mediterranean European countries, with extensive assessment at baseline and after one-year of intervention; (2) a well-controlled nutritional intervention; (3) the combination of data on genetics and epigenetics for the same individuals; and (4) the use of the state-of-the-art epigenetic biomarkers of aging, which are robust and well-validated instruments, previously associated with mortality, morbidity, and several age-related phenotypes (Horvath and Raj 2018).
On the contrary, several important limitations exist. Firstly, the absence of a control group is a major one. Genome-wide DNA methylation analysis was performed only in a subset of subjects included in the intervention group and was not performed on subjects randomized into the control group. This outline corresponds to the initial design of the NU-AGE study, as described by Santoro et al. (Santoro et al. 2014), where it was originally decided to perform OMICs analysis (epigenomics, transcriptomics, metabolomics, and metagenomics) in a subgroup of 120 randomly selected subjects before and after diet. There were no specific changes in the lifestyle behaviors of the individuals recruited in the control group, and specially, no significant increase in the NU-AGE index, which measures the adherence to Mediterranean diet. This index, while not different between the two groups at baseline, only changed significantly in the intervention group after 1 year, as compared with the control one (Berendsen et al. 2018). In support of our data, it has been demonstrated that Mediterranean diet can have a specific impact on DNA methylation levels of certain genes. Thus, in the PREDIMED study, it was observed that a nutritional intervention based on Mediterranean diet was able to impact on DNA methylation levels of inflammation-related genes compared with the non-intervention group (Arpón et al. 2017).
Secondly, in our experimental settings, the nutritional intervention resulted only in a small effect size (Supplementary Table 1), which however is reasonable according to recent literature. In the recently published results of the TRIIM trial (Fahy et al. 2019), the nine volunteers were found to be on average 2.5 years younger after the intervention than they entered, which means that they gained back about 1.5 years in 1 year of trial. It is reasonable that the effect of NU-AGE nutritional intervention (i.e., average of − 1.47 years of AgeAccelDiff and of − 1.36 years of IEAADiff in the subgroup of Polish females) could have a minor effect on aging biomarkers compared with the one presented by Fahy et al., who evaluated the impact of 3 combined drugs. However, this small effect should be necessarily confirmed in a larger number of enrolled subjects compared with the cohort analyzed in our pilot study. The same applies to our GWAS analysis, as no significant association was observed after correction for multiple testing and as the small sample size prevented us from performing the analysis in subgroups divided for country or sex.
In conclusion, we report that a Mediterranean-like nutritional intervention can promote epigenetic rejuvenation in the elderly, and that its effect is dependent on different factors including the following: (1) country-/population-specific factors, likely influenced by anthropologic and cultural components; (2) sex-/gender-specific factors: and (3) individual-specific factors, for example, related to the genetic background and to the baseline epigenetic profile of each individual.
Further work is required to overcome the important limitations of this preliminary work, to elucidate how some specific determinants influence the epigenetic aging and how some individuals seem to be more prone to benefit from specific interventions. This will be a key achievement for the development of individualized nutritional interventions aimed at promoting healthy living and, more in general, for the application of a precision medicine approach to anti-aging interventions.
References
Ambatipudi S, Horvath S, Perrier F, Cuenin C, Hernandez-Vargas H, le Calvez-Kelm F, Durand G, Byrnes G, Ferrari P, Bouaoun L, Sklias A, Chajes V, Overvad K, Severi G, Baglietto L, Clavel-Chapelon F, Kaaks R, Barrdahl M, Boeing H, Trichopoulou A, Lagiou P, Naska A, Masala G, Agnoli C, Polidoro S, Tumino R, Panico S, Dollé M, Peeters PHM, Onland-Moret NC, Sandanger TM, Nøst TH, Weiderpass E, Quirós JR, Agudo A, Rodriguez-Barranco M, Huerta Castaño JM, Barricarte A, Fernández AM, Travis RC, Vineis P, Muller DC, Riboli E, Gunter M, Romieu I, Herceg Z (2017) DNA methylome analysis identifies accelerated epigenetic ageing associated with postmenopausal breast cancer susceptibility. Eur J Cancer 75:299–307. https://doi.org/10.1016/j.ejca.2017.01.014
Arpón A, Riezu-Boj JI, Milagro FI, Marti A, Razquin C, Martínez-González MA, Corella D, Estruch R, Casas R, Fitó M, Ros E, Salas-Salvadó J, Martínez JA (2017) Adherence to Mediterranean diet is associated with methylation changes in inflammation-related genes in peripheral blood cells. J Physiol Biochem 73:445–455. https://doi.org/10.1007/s13105-017-0552-6
Athanasopoulou S, Chondrogianni N, Santoro A, Asimaki K, Delitsikou V, Voutetakis K, Fabbri C, Pietruszka B, Kaluza J, Franceschi C, Gonos ES (2018) Beneficial effects of elderly tailored Mediterranean diet on the proteasomal proteolysis. Front Physiol 9:457. https://doi.org/10.3389/fphys.2018.00457
Bacalini MG, Friso S, Olivieri F, Pirazzini C, Giuliani C, Capri M, Santoro A, Franceschi C, Garagnani P (2014) Present and future of anti-ageing epigenetic diets. Mech Ageing Dev 136–137:101–115. https://doi.org/10.1016/j.mad.2013.12.006
Berendsen A, Santoro A, Pini E, Cevenini E, Ostan R, Pietruszka B, Rolf K, Cano N, Caille A, Lyon-Belgy N, Fairweather-Tait S, Feskens E, Franceschi C, de Groot CP (2014) Reprint of: a parallel randomized trial on the effect of a healthful diet on inflammageing and its consequences in European elderly people: design of the NU-AGE dietary intervention study. Mech Ageing Dev 136–137:14–21. https://doi.org/10.1016/j.mad.2014.03.001
Berendsen AAM, van de Rest O, Feskens EJM et al (2018) Changes in dietary intake and adherence to the NU-AGE diet following a one-year dietary intervention among European older adults-results of the NU-AGE randomized trial. Nutrients:10. https://doi.org/10.3390/nu10121905
Breitling LP, Saum K-U, Perna L, Schöttker B, Holleczek B, Brenner H (2016) Frailty is associated with the epigenetic clock but not with telomere length in a German cohort. Clin Epigenetics 8:21. https://doi.org/10.1186/s13148-016-0186-5
Chen BH, Marioni RE, Colicino E et al (2016) DNA methylation-based measures of biological age: meta-analysis predicting time to death. Aging (Albany NY) 8:1844–1865. https://doi.org/10.18632/aging.101020
Christiansen L, Lenart A, Tan Q, Vaupel JW, Aviv A, McGue M, Christensen K (2016) DNA methylation age is associated with mortality in a longitudinal Danish twin study. Aging Cell 15:149–154. https://doi.org/10.1111/acel.12421
Cummings NE, Lamming DW (2017) Regulation of metabolic health and aging by nutrient-sensitive signaling pathways. Mol Cell Endocrinol 455:13–22. https://doi.org/10.1016/j.mce.2016.11.014
Dato S, Bellizzi D, Rose G, Passarino G (2016) The impact of nutrients on the aging rate: a complex interaction of demographic, environmental and genetic factors. Mech Ageing Dev 154:49–61. https://doi.org/10.1016/j.mad.2016.02.005
Davinelli S, Trichopoulou A, Corbi G, de Vivo I, Scapagnini G (2019) The potential nutrigeroprotective role of Mediterranean diet and its functional components on telomere length dynamics. Ageing Res Rev 49:1–10. https://doi.org/10.1016/j.arr.2018.11.001
Degerman S, Josefsson M, Nordin Adolfsson A, Wennstedt S, Landfors M, Haider Z, Pudas S, Hultdin M, Nyberg L, Adolfsson R (2017) Maintained memory in aging is associated with young epigenetic age. Neurobiol Aging 55:167–171. https://doi.org/10.1016/j.neurobiolaging.2017.02.009
Dugué P-A, Bassett JK, Joo JE et al (2018) Association of DNA methylation-based biological age with health risk factors and overall and cause-specific mortality. Am J Epidemiol 187:529–538. https://doi.org/10.1093/aje/kwx291
Estruch R, Martínez-González MA, Corella D, Salas-Salvadó J, Ruiz-Gutiérrez V, Covas MI, Fiol M, Gómez-Gracia E, López-Sabater MC, Vinyoles E, Arós F, Conde M, Lahoz C, Lapetra J, Sáez G, Ros E, PREDIMED Study Investigators (2006) Effects of a Mediterranean-style diet on cardiovascular risk factors: a randomized trial. Ann Intern Med 145:1–11
Estruch R, Ros E, Salas-Salvadó J, Covas MI, Corella D, Arós F, Gómez-Gracia E, Ruiz-Gutiérrez V, Fiol M, Lapetra J, Lamuela-Raventos RM, Serra-Majem L, Pintó X, Basora J, Muñoz MA, Sorlí JV, Martínez JA, Martínez-González MA, PREDIMED Study Investigators (2013) Primary prevention of cardiovascular disease with a Mediterranean diet. N Engl J Med 368:1279–1290. https://doi.org/10.1056/NEJMoa1200303
Fahy GM, Brooke RT, Watson JP et al (2019) Reversal of epigenetic aging and immunosenescent trends in humans. Aging Cell:e13028. https://doi.org/10.1111/acel.13028
Franceschi C, Bonafè M, Valensin S, Olivieri F, de Luca M, Ottaviani E, de Benedictis G (2000) Inflamm-aging. An evolutionary perspective on immunosenescence. Ann N Y Acad Sci 908:244–254
Fried LP, Tangen CM, Walston J et al (2001) Frailty in older adults: evidence for a phenotype. J Gerontol A Biol Sci Med Sci 56:M146–M156
Gale CR, Marioni RE, Čukić I, Chastin SF, Dall PM, Dontje ML, Skelton DA, Deary IJ, Seniors USP Team (2018a) The epigenetic clock and objectively measured sedentary and walking behavior in older adults: the Lothian Birth Cohort 1936. Clin Epigenetics 10:4. https://doi.org/10.1186/s13148-017-0438-z
Gale CR, Marioni RE, Harris SE, Starr JM, Deary IJ (2018b) DNA methylation and the epigenetic clock in relation to physical frailty in older people: the Lothian Birth Cohort 1936. Clin Epigenetics 10:101. https://doi.org/10.1186/s13148-018-0538-4
Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, Klotzle B, Bibikova M, Fan JB, Gao Y, Deconde R, Chen M, Rajapakse I, Friend S, Ideker T, Zhang K (2013) Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell 49:359–367. https://doi.org/10.1016/j.molcel.2012.10.016
Heiss C, Spyridopoulos I, Haendeler J (2017) Interventions to slow cardiovascular aging: dietary restriction, drugs and novel molecules. Exp Gerontol. https://doi.org/10.1016/j.exger.2017.06.015
Horvath S (2013) DNA methylation age of human tissues and cell types. Genome Biol 14:R115. https://doi.org/10.1186/gb-2013-14-10-r115
Horvath S, Erhart W, Brosch M, Ammerpohl O, von Schönfels W, Ahrens M, Heits N, Bell JT, Tsai PC, Spector TD, Deloukas P, Siebert R, Sipos B, Becker T, Röcken C, Schafmayer C, Hampe J (2014) Obesity accelerates epigenetic aging of human liver. Proc Natl Acad Sci U S A 111:15538–15543. https://doi.org/10.1073/pnas.1412759111
Horvath S, Gurven M, Levine ME et al (2016) An epigenetic clock analysis of race/ethnicity, sex, and coronary heart disease. Genome Biol 17:171. https://doi.org/10.1186/s13059-016-1030-0
Horvath S, Pirazzini C, Bacalini MG et al (2015) Decreased epigenetic age of PBMCs from Italian semi-supercentenarians and their offspring. Aging (Albany NY) 7:1159–1170. https://doi.org/10.18632/aging.100861
Horvath S, Raj K (2018) DNA methylation-based biomarkers and the epigenetic clock theory of ageing. Nat Rev Genet 19:371–384. https://doi.org/10.1038/s41576-018-0004-3
Horvath S, Ritz BR (2015) Increased epigenetic age and granulocyte counts in the blood of Parkinson’s disease patients. Aging (Albany NY) 7:1130–1142. https://doi.org/10.18632/aging.100859
Houseman EA, Accomando WP, Koestler DC, Christensen BC, Marsit CJ, Nelson HH, Wiencke JK, Kelsey KT (2012) DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics 13:86. https://doi.org/10.1186/1471-2105-13-86
Jennings A, Berendsen AM, de Groot LCPGM, et al (2019) Mediterranean-style diet improves systolic blood pressure and arterial stiffness in older adults. Hypertension HYPERTENSIONAHA 11812259. Doi: https://doi.org/10.1161/HYPERTENSIONAHA.118.12259
Jennings A, Cashman KD, Gillings R et al (2018) A Mediterranean-like dietary pattern with vitamin D3 (10 μg/d) supplements reduced the rate of bone loss in older Europeans with osteoporosis at baseline: results of a 1-y randomized controlled trial. Am J Clin Nutr. https://doi.org/10.1093/ajcn/nqy122
Kojima G, Avgerinou C, Iliffe S, Walters K (2018) Adherence to Mediterranean diet reduces incident frailty risk: systematic review and meta-analysis. J Am Geriatr Soc. https://doi.org/10.1111/jgs.15251
Kok DEG, Dhonukshe-Rutten RAM, Lute C, Heil SG, Uitterlinden AG, van der Velde N, van Meurs J, van Schoor N, Hooiveld GJ, de Groot LC, Kampman E, Steegenga WT (2015) The effects of long-term daily folic acid and vitamin B12 supplementation on genome-wide DNA methylation in elderly subjects. Clin Epigenetics 7:121. https://doi.org/10.1186/s13148-015-0154-5
Konz T, Santoro A, Goulet L, Bazzocchi A, Battista G, Nicoletti C, Kadi F, Ostan R, Goy M, Monnard C, Martin FP, Feige JN, Franceschi C, Rezzi S (2018) Sex-specific associations of blood-based nutrient profiling with body composition in the elderly. Front Physiol 9:1935. https://doi.org/10.3389/fphys.2018.01935
Levine ME, Hosgood HD, Chen B et al (2015) DNA methylation age of blood predicts future onset of lung cancer in the women’s health initiative. Aging (Albany NY) 7:690–700. https://doi.org/10.18632/aging.100809
Levine ME, Lu AT, Quach A et al (2018) An epigenetic biomarker of aging for lifespan and healthspan. Aging (Albany NY) 10:573–591. https://doi.org/10.18632/aging.101414
Li J, Zhu X, Yu K, Jiang H, Zhang Y, Wang B, Liu X, Deng S, Hu J, Deng Q, Sun H, Guo H, Zhang X, Chen W, Yuan J, He M, Bai Y, Han X, Liu B, Liu C, Guo Y, Zhang B, Zhang Z, Hu FB, Gao W, Li L, Lathrop M, Laprise C, Liang L, Wu T (2018) Exposure to polycyclic aromatic hydrocarbons and accelerated DNA methylation aging. Environ Health Perspect 126:067005. https://doi.org/10.1289/EHP2773
Liao C-Y, Rikke BA, Johnson TE, Diaz V, Nelson JF (2010) Genetic variation in the murine lifespan response to dietary restriction: from life extension to life shortening. Aging Cell 9:92–95. https://doi.org/10.1111/j.1474-9726.2009.00533.x
Longo VD, Antebi A, Bartke A, Barzilai N, Brown-Borg HM, Caruso C, Curiel TJ, de Cabo R, Franceschi C, Gems D, Ingram DK, Johnson TE, Kennedy BK, Kenyon C, Klein S, Kopchick JJ, Lepperdinger G, Madeo F, Mirisola MG, Mitchell JR, Passarino G, Rudolph KL, Sedivy JM, Shadel GS, Sinclair DA, Spindler SR, Suh Y, Vijg J, Vinciguerra M, Fontana L (2015) Interventions to slow aging in humans: are we ready? Aging Cell 14:497–510. https://doi.org/10.1111/acel.12338
Lu AT, Hannon E, Levine ME, Hao K, Crimmins EM, Lunnon K, Kozlenkov A, Mill J, Dracheva S, Horvath S (2016) Genetic variants near MLST8 and DHX57 affect the epigenetic age of the cerebellum. Nat Commun 7:10561. https://doi.org/10.1038/ncomms10561
Lu AT, Hannon E, Levine ME, Crimmins EM, Lunnon K, Mill J, Geschwind DH, Horvath S (2017) Genetic architecture of epigenetic and neuronal ageing rates in human brain regions. Nat Commun 8:15353. https://doi.org/10.1038/ncomms15353
Lu AT, Quach A, Wilson JG et al (2019) DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging (Albany NY) 11:303–327. https://doi.org/10.18632/aging.101684
Lu AT, Xue L, Salfati EL, Chen BH, Ferrucci L, Levy D, Joehanes R, Murabito JM, Kiel DP, Tsai PC, Yet I, Bell JT, Mangino M, Tanaka T, McRae A, Marioni RE, Visscher PM, Wray NR, Deary IJ, Levine ME, Quach A, Assimes T, Tsao PS, Absher D, Stewart JD, Li Y, Reiner AP, Hou L, Baccarelli AA, Whitsel EA, Aviv A, Cardona A, Day FR, Wareham NJ, Perry JRB, Ong KK, Raj K, Lunetta KL, Horvath S (2018) GWAS of epigenetic aging rates in blood reveals a critical role for TERT. Nat Commun 9:387. https://doi.org/10.1038/s41467-017-02697-5
Lushchak O, Strilbytska OM, Yurkevych I, Vaiserman AM, Storey KB (2019) Implications of amino acid sensing and dietary protein to the aging process. Exp Gerontol 115:69–78. https://doi.org/10.1016/j.exger.2018.11.021
Maierhofer A, Flunkert J, Oshima J et al (2017) Accelerated epigenetic aging in Werner syndrome. Aging (Albany NY) 9:1143–1152. https://doi.org/10.18632/aging.101217
Maijo M, Ivory K, Clements SJ, Dainty JR, Jennings A, Gillings R, Fairweather-Tait S, Gulisano M, Santoro A, Franceschi C, Carding SR, Nicoletti C (2018) One-year consumption of a Mediterranean-like dietary pattern with vitamin D3 supplements induced small scale but extensive changes of immune cell phenotype, co-receptor expression and innate immune responses in healthy elderly subjects: results from the United Kingdom arm of the NU-AGE trial. Front Physiol 9:997. https://doi.org/10.3389/fphys.2018.00997
Marioni RE, Shah S, McRae AF, Chen BH, Colicino E, Harris SE, Gibson J, Henders AK, Redmond P, Cox SR, Pattie A, Corley J, Murphy L, Martin NG, Montgomery GW, Feinberg AP, Fallin MD, Multhaup ML, Jaffe AE, Joehanes R, Schwartz J, Just AC, Lunetta KL, Murabito JM, Starr JM, Horvath S, Baccarelli AA, Levy D, Visscher PM, Wray NR, Deary IJ (2015a) DNA methylation age of blood predicts all-cause mortality in later life. Genome Biol 16:25. https://doi.org/10.1186/s13059-015-0584-6
Marioni RE, Shah S, McRae AF, Ritchie SJ, Muniz-Terrera G, Harris SE, Gibson J, Redmond P, Cox SR, Pattie A, Corley J, Taylor A, Murphy L, Starr JM, Horvath S, Visscher PM, Wray NR, Deary IJ (2015b) The epigenetic clock is correlated with physical and cognitive fitness in the Lothian Birth Cohort 1936. Int J Epidemiol 44:1388–1396. https://doi.org/10.1093/ije/dyu277
Marseglia A, Xu W, Fratiglioni L, Fabbri C, Berendsen AAM, Bialecka-Debek A, Jennings A, Gillings R, Meunier N, Caumon E, Fairweather-Tait S, Pietruszka B, de Groot LCPGM, Santoro A, Franceschi C (2018) Effect of the NU-AGE diet on cognitive functioning in older adults: a randomized controlled trial. Front Physiol 9:349. https://doi.org/10.3389/fphys.2018.00349
Martínez-González MÁ, Hershey MS, Zazpe I, Trichopoulou A (2017) Transferability of the Mediterranean diet to non-Mediterranean countries. What Is and What Is Not the Mediterranean Diet. Nutrients 9. https://doi.org/10.3390/nu9111226
Martínez-González MA, Salas-Salvadó J, Estruch R, Corella D, Fitó M, Ros E, PREDIMED INVESTIGATORS (2015) Benefits of the Mediterranean diet: insights from the PREDIMED study. Prog Cardiovasc Dis 58:50–60. https://doi.org/10.1016/j.pcad.2015.04.003
Martucci M, Ostan R, Biondi F, Bellavista E, Fabbri C, Bertarelli C, Salvioli S, Capri M, Franceschi C, Santoro A (2017) Mediterranean diet and inflammaging within the hormesis paradigm. Nutr Rev 75:442–455. https://doi.org/10.1093/nutrit/nux013
Mayr HL, Tierney AC, Thomas CJ, Ruiz-Canela M, Radcliffe J, Itsiopoulos C (2018) Mediterranean-type diets and inflammatory markers in patients with coronary heart disease: a systematic review and meta-analysis. Nutr Res 50:10–24. https://doi.org/10.1016/j.nutres.2017.10.014
Mitjavila MT, Fandos M, Salas-Salvadó J et al (2013) The Mediterranean diet improves the systemic lipid and DNA oxidative damage in metabolic syndrome individuals. A randomized, controlled, trial. Clin Nutr 32:172–178. https://doi.org/10.1016/j.clnu.2012.08.002
Nevalainen T, Kananen L, Marttila S, Jylhävä J, Mononen N, Kähönen M, Raitakari OT, Hervonen A, Jylhä M, Lehtimäki T, Hurme M (2017) Obesity accelerates epigenetic aging in middle-aged but not in elderly individuals. Clin Epigenetics 9:20. https://doi.org/10.1186/s13148-016-0301-7
Ostan R, Guidarelli G, Giampieri E, Lanzarini C, Berendsen AAM, Januszko O, Jennings A, Lyon N, Caumon E, Gillings R, Sicinska E, Meunier N, Feskens EJM, Pietruszka B, de Groot LCPGM, Fairweather-Tait S, Capri M, Franceschi C, Santoro A (2018) Cross-sectional analysis of the correlation between daily nutrient intake assessed by 7-day food records and biomarkers of dietary intake among participants of the NU-AGE study. Front Physiol 9:1359. https://doi.org/10.3389/fphys.2018.01359
Ostan R, Lanzarini C, Pini E et al (2015) Inflammaging and cancer: a challenge for the Mediterranean diet. Nutrients 7:2589–2621. https://doi.org/10.3390/nu7042589
Pavanello S, Campisi M, Tona F et al (2019) Exploring epigenetic age in response to intensive relaxing training: a pilot study to slow down biological age. Int J Environ Res Public Health 16. https://doi.org/10.3390/ijerph16173074
Perna L, Zhang Y, Mons U, Holleczek B, Saum KU, Brenner H (2016) Epigenetic age acceleration predicts cancer, cardiovascular, and all-cause mortality in a German case cohort. Clin Epigenetics 8:64. https://doi.org/10.1186/s13148-016-0228-z
Pujos-Guillot E, Pétéra M, Jacquemin J, Centeno D, Lyan B, Montoliu I, Madej D, Pietruszka B, Fabbri C, Santoro A, Brzozowska A, Franceschi C, Comte B (2018) Identification of pre-frailty sub-phenotypes in elderly using metabolomics. Front Physiol 9:1903. https://doi.org/10.3389/fphys.2018.01903
Quach A, Levine ME, Tanaka T et al (2017) Epigenetic clock analysis of diet, exercise, education, and lifestyle factors. Aging (Albany NY) 9:419–446. https://doi.org/10.18632/aging.101168
Rosen AD, Robertson KD, Hlady RA, Muench C, Lee J, Philibert R, Horvath S, Kaminsky ZA, Lohoff FW (2018) DNA methylation age is accelerated in alcohol dependence. Transl Psychiatry 8:182. https://doi.org/10.1038/s41398-018-0233-4
Sae-Lee C, Corsi S, Barrow TM et al (2018) Dietary intervention modifies DNA methylation age assessed by the epigenetic clock. Mol Nutr Food Res:e1800092. https://doi.org/10.1002/mnfr.201800092
Santoro A, Bazzocchi A, Guidarelli G, Ostan R, Giampieri E, Mercatelli D, Scurti M, Berendsen A, Surala O, Jennings A, Meunier N, Caumon E, Gillings R, Kadi F, Capel F, Cashman KD, Pietruszka B, Feskens EJM, de Groot LCPGM, Battista G, Salvioli S, Franceschi C (2018) A cross-sectional analysis of body composition among healthy elderly from the European NU-AGE study: sex and country specific features. Front Physiol 9:1693. https://doi.org/10.3389/fphys.2018.01693
Santoro A, Guidarelli G, Ostan R, Giampieri E, Fabbri C, Bertarelli C, Nicoletti C, Kadi F, de Groot LCPGM, Feskens E, Berendsen A, Brzozowska A, Januszko O, Kozlowska K, Fairweather-Tait S, Jennings A, Meunier N, Caumon E, Napoli A, Mercatelli D, Battista G, Capri M, Franceschi C, Bazzocchi A (2019) Gender-specific association of body composition with inflammatory and adipose-related markers in healthy elderly Europeans from the NU-AGE study. Eur Radiol 29:4968–4979. https://doi.org/10.1007/s00330-018-5973-2
Santoro A, Pini E, Scurti M et al (2014) Combating inflammaging through a Mediterranean whole diet approach: the NU-AGE project’s conceptual framework and design. Mech Ageing Dev 136–137:3–13. https://doi.org/10.1016/j.mad.2013.12.001
Shlisky J, Bloom DE, Beaudreault AR, Tucker KL, Keller HH, Freund-Levi Y, Fielding RA, Cheng FW, Jensen GL, Wu D, Meydani SN (2017) Nutritional considerations for healthy aging and reduction in age-related chronic disease. Adv Nutr 8:17–26. https://doi.org/10.3945/an.116.013474
Sillanpää E, Laakkonen EK, Vaara E, Rantanen T, Kovanen V, Sipilä S, Kaprio J, Ollikainen M (2018) Biological clocks and physical functioning in monozygotic female twins. BMC Geriatr 18:83. https://doi.org/10.1186/s12877-018-0775-6
Simpkin AJ, Cooper R, Howe LD, Relton CL, Davey Smith G, Teschendorff A, Widschwendter M, Wong A, Kuh D, Hardy R (2017) Are objective measures of physical capability related to accelerated epigenetic age? Findings from a British birth cohort. BMJ Open 7:e016708. https://doi.org/10.1136/bmjopen-2017-016708
Tosti V, Bertozzi B, Fontana L (2018) Health benefits of the Mediterranean diet: metabolic and molecular mechanisms. J Gerontol A Biol Sci Med Sci 73:318–326. https://doi.org/10.1093/gerona/glx227
Touleimat N, Tost J (2012) Complete pipeline for Infinium(®) human methylation 450K BeadChip data processing using subset quantile normalization for accurate DNA methylation estimation. Epigenomics 4:325–341. https://doi.org/10.2217/epi.12.21
Wahl D, Cogger VC, Solon-Biet SM, Waern RV, Gokarn R, Pulpitel T, Cabo Rd, Mattson MP, Raubenheimer D, Simpson SJ, le Couteur DG (2016) Nutritional strategies to optimise cognitive function in the aging brain. Ageing Res Rev 31:80–92. https://doi.org/10.1016/j.arr.2016.06.006
Weidner CI, Lin Q, Koch CM, Eisele L, Beier F, Ziegler P, Bauerschlag DO, Jöckel KH, Erbel R, Mühleisen TW, Zenke M, Brümmendorf TH, Wagner W (2014) Aging of blood can be tracked by DNA methylation changes at just three CpG sites. Genome Biol 15:R24. https://doi.org/10.1186/gb-2014-15-2-r24
WHO | Global health and ageing [Internet] WHO (cited 2017). Available from: http://www.who.int/ageing/publications/global_health/en/
Xia Z, Cholewa J, Zhao Y, Shang HY, Yang YQ, Araújo Pessôa K, Su QS, Lima-Soares F, Zanchi NE (2017) Targeting inflammation and downstream protein metabolism in sarcopenia: a brief up-dated description of concurrent exercise and leucine-based multimodal intervention. Front Physiol 8:434. https://doi.org/10.3389/fphys.2017.00434
Xiao F-H, Chen X-Q, He Y-H, Kong Q-P (2018) Accelerated DNA methylation changes in middle-aged men define sexual dimorphism in human lifespans. Clin Epigenetics 10:133. https://doi.org/10.1186/s13148-018-0573-1
Zbieć-Piekarska R, Spólnicka M, Kupiec T, Parys-Proszek A, Makowska Ż, Pałeczka A, Kucharczyk K, Płoski R, Branicki W (2015) Development of a forensically useful age prediction method based on DNA methylation analysis. Forensic Sci Int Genet 17:173–179. https://doi.org/10.1016/j.fsigen.2015.05.001
Acknowledgments
We thank Maria Scurti, Claudia Bertarelli, Maria Giustina Palmas, Massimo Izzi, Elisa Pini and Catia Lanzarini for their technical support.
Funding
This study was supported by the European Union’s Seventh Framework Program under grant agreement n°266486 (“NU-AGE: New dietary strategies addressing the specific needs of the elderly population for healthy aging in Europe”) and by the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement n°675003 (“PANINI: Physical Activity and Nutrition Influences In ageing”). We acknowledge support by the grant of the Ministry of Education and Science of the Russian Federation Agreement No. 075-15-2019-871.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Gensous, N., Garagnani, P., Santoro, A. et al. One-year Mediterranean diet promotes epigenetic rejuvenation with country- and sex-specific effects: a pilot study from the NU-AGE project. GeroScience 42, 687–701 (2020). https://doi.org/10.1007/s11357-019-00149-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11357-019-00149-0