Abstract
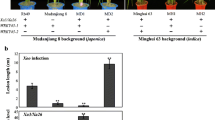
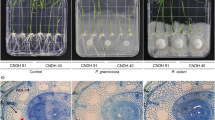
Bacterial blight caused by Xanthomonas oryzae pv. oryzae (Xoo) is the most harmful bacterial disease of rice worldwide. Previously, we characterized major disease resistance (MR) gene xa25, which confers race-specific resistance to Xoo strain PXO339. The xa25 is a recessive allele of the SWEET13 locus, but SWEET13’s interaction with PXO339 and how efficiently using this locus for rice breeding still need to be defined. Here we show that the SWEET13 allele from rice Zhenshan 97 is a susceptibility gene to PXO339. Using this allele’s promoter to regulate xa25 resulted in disease, suggesting that the promoter is a key determinant in SWEET13 caused disease in Zhanshan 97 after PXO339 infection. PXO339 transcriptionally induces SWEET13 to cause disease. Partial suppressing SWEET13 expression leads to a high level of resistance to PXO339. Thus, the transcriptionally suppressed SWEET13 functions as xa25 in resistance to PXO339. Hybrid rice is widely grown in many countries. However, recessive MR genes have not been efficiently used for disease resistance breeding in hybrid rice production for both parents of the hybrid have to carry the same recessive gene. However, the suppressed SWEET13 functions dominantly, which will have advantage to improve the resistance of hybrid rice to xa25-incomptible Xoo.
Similar content being viewed by others
References
Antony, G., Zhou, J., Huang, S., Li, T., Liu, B., White, F., and Yang, B. (2010). Rice xa13 recessive resistance to bacterial blight is defeated by induction of the disease susceptibility gene Os-11N3. Plant Cell 22, 3864–3876.
Boch, J., Scholze, H., Schornack, S., Landgraf, A., Hahn, S., Kay, S., Lahaye, T., Nickstadt, A., and Bonas, U. (2009). Breaking the code of DNA binding specificity of TAL-Type III effectors. Science 326, 1509–1512.
Cao, Y., Duan, L., Li, H., Sun, X., Zhao, Y., Xu, C., Li, X., and Wang, S. (2007). Functional analysis of Xa3/Xa26 family members in rice resistance to Xanthomonas oryzae pv. oryzae. Theor Appl Genet 115, 887–895.
Chardon, F., Bedu, M., Calenge, F., Klemens, P.A.W., Spinner, L., Clement, G., Chietera, G., Léran, S., Ferrand, M., Lacombe, B., Loudet, O., Dinant, S., Bellini, C., Neuhaus, H.E., Daniel-Vedele, F., and Krapp, A. (2013). Leaf fructose content is controlled by the vacuolar transporter SWEET17 in Arabidopsis. Curr Biol 23, 697–702.
Chen, H., Wang, S., and Zhang, Q. (2002). New gene for bacterial blight resistance in rice located on chromosome 12 identified from Minghui 63, an elite restorer line. Phytopathology 92, 750–754.
Chen, L.Q., Hou, B.H., Lalonde, S., Takanaga, H., Hartung, M.L., Qu, X.Q., Guo, W.J., Kim, J.G., Underwood, W., Chaudhuri, B., Chermak, D., Antony, G., White, F.F., Somerville, S.C., Mudgett, M.B., and Frommer, W.B. (2010). Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 468, 527–532.
Chu, Z., Yuan, M., Yao, J., Ge, X., Yuan, B., Xu, C., Li, X., Fu, B., Li, Z., Bennetzen, J.L., Zhang, Q., and Wang, S. (2006). Promoter mutations of an essential gene for pollen development result in disease resistance in rice. Genes Dev 20, 1250–1255.
Ge, X., Chu, Z., Lin, Y., and Wang, S. (2006). A tissue culture system for different germplasms of indica rice. Plant Cell Rep 25, 392–402.
Hutin, M., Sabot, F., Ghesquiè re, A., Koebnik, R., and Szurek, B. (2015). A knowledge-based molecular screen uncovers a broad-spectrum OsSWEET14 resistance allele to bacterial blight from wild rice. Plant J 84, 694–703.
Ke, Y., Liu, H., Li, X., Xiao, J., and Wang, S. (2014). Rice Os PAD4 functions differently from Arabidopsis at PAD4 in host-pathogen interactions. Plant J 78, 619–631.
Kou, Y., and Wang, S. (2013). Bacterial blight resistance in rice. In Translational Genomics for Crop Breeding: Volume 1—Biotic Stress, R.K. Varshney, and R. Tuberosa, eds. (Hoboken: Wiley-Blackwell/John Wiley & Sons, Inc.), pp. 11–30.
Liu, Q., Yuan, M., Zhou, Y., Li, X., Xiao, J., and Wang, S. (2011). A paralog of the MtN3/saliva family recessively confers race-specific resistance to Xanthomonas oryzae in rice. Plant Cell Environ 34, 1958–1969.
Moscou, M.J., and Bogdanove, A.J. (2009). A simple cipher governs DNA recognition by TAL effectors. Science 326, 1501–1501.
Qiu, D., Xiao, J., Ding, X., Xiong, M., Cai, M., Cao, Y., Li, X., Xu, C., and Wang, S. (2007). OsWRKY13 mediates rice disease resistance by regulating defense-related genes in salicylate- and jasmonate-dependent signaling. Mol Plant Microbe Interact 20, 492–499.
Richter, A., Streubel, J., Blücher, C., Szurek, B., Reschke, M., Grau, J., and Boch, J. (2014). A TAL effector repeat architecture for frameshift binding. Nat Commun 5, 3447.
Schornack, S., Moscou, M.J., Ward, E.R., and Horvath, D.M. (2013). Engineering plant disease resistance based on TAL effectors. Annu Rev Phytopathol 51, 383–406.
Wang, S., Sun, J., Fan, F., Tan, Z., Zou, Y., and Lu, D. (2016). A Xanthomonas oryzae pv. oryzae effector, XopR, associates with receptor-like cytoplasmic kinases and suppresses PAMP-triggered stomatal closure. Sci China Life Sci 59, 897–905.
Warrens, A.N., Jones, M.D., and Lechler, R.I. (1997). Splicing by overlap extension by PCR using asymmetric amplification: an improved technique for the generation of hybrid proteins of immunological interest. Gene 186, 29–35.
Warthmann, N., Chen, H., Ossowski, S., Weigel, D., and Hervé, P. (2008). Highly specific gene silencing by artificial miRNAs in rice. PLoS ONE 3, e1829.
Wen, N., Chu, Z., and Wang, S. (2003). Three types of defense-responsive genes are involved in resistance to bacterial blight and fungal blast diseases in rice. Mol Genets Genomics 269, 331–339.
Wu, C., Li, X., Yuan, W., Chen, G., Kilian, A., Li, J., Xu, C., Li, X., Zhou, D.X., Wang, S., and Zhang, Q. (2003). Development of enhancer trap lines for functional analysis of the rice genome. Plant J 35, 418–427.
Yang, B., Sugio, A., and White, F.F. (2006). Os8N3 is a host disease-susceptibility gene for bacterial blight of rice. Proc Natl Acad Sci USA 103, 10503–10508.
Yuan, M., Chu, Z., Li, X., Xu, C., and Wang, S. (2009). Pathogen-induced expressional loss of function is the key factor in race-specific bacterial resistance conferred by a recessive R gene xa13 in rice. Plant Cell Physiol 50, 947–955.
Yuan, M., Ke, Y., Huang, R., Ma, L., Yang, Z., Chu, Z., Xiao, J., Li, X., and Wang, S. (2016). A host basal transcription factor is a key component for infection of rice by TALE-carrying bacteria. eLife 5, e19605.
Yuan, M., and Wang, S. (2013). Rice MtN3/Saliva/SWEET family genes and their homologs in cellular organisms. Mol Plant 6, 665–674.
Yuan, M., Zhao, J., Huang, R., Li, X., Xiao, J., and Wang, S. (2014). Rice MtN3/saliva/SWEET gene family: evolution, expression profiling, and sugar transport. J Integr Plant Biol 56, 559–570.
Zhang, H., and Wang, S. (2013). Rice versus Xanthomonas oryzae pv. oryzae: a unique pathosystem. Curr Opin Plant Biol 16, 188–195.
Zhou, J., Peng, Z., Long, J., Sosso, D., Liu, B., Eom, J.S., Huang, S., Liu, S., Vera Cruz, C., Frommer, W.B., White, F.F., and Yang, B. (2015). Gene targeting by the TAL effector PthXo2 reveals cryptic resistance gene for bacterial blight of rice. Plant J 82, 632–643.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (31330062), and the National Key Research and Development Program of Chin (2016YFD0100903).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
11427_2016_299_MOESM1_ESM.pdf
Characterization of a disease susceptibility locus for exploring an efficient way to improve rice resistance against bacterial blight
Rights and permissions
About this article
Cite this article
Cheng, Q., Mao, W., Xie, W. et al. Characterization of a disease susceptibility locus for exploring an efficient way to improve rice resistance against bacterial blight. Sci. China Life Sci. 60, 298–306 (2017). https://doi.org/10.1007/s11427-016-0299-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11427-016-0299-x