Abstract
Purpose
Surgical data science is a new research field that aims to observe all aspects of the patient treatment process in order to provide the right assistance at the right time. Due to the breakthrough successes of deep learning-based solutions for automatic image annotation, the availability of reference annotations for algorithm training is becoming a major bottleneck in the field. The purpose of this paper was to investigate the concept of self-supervised learning to address this issue.
Methods
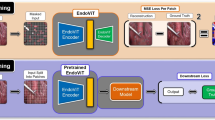
Our approach is guided by the hypothesis that unlabeled video data can be used to learn a representation of the target domain that boosts the performance of state-of-the-art machine learning algorithms when used for pre-training. Core of the method is an auxiliary task based on raw endoscopic video data of the target domain that is used to initialize the convolutional neural network (CNN) for the target task. In this paper, we propose the re-colorization of medical images with a conditional generative adversarial network (cGAN)-based architecture as auxiliary task. A variant of the method involves a second pre-training step based on labeled data for the target task from a related domain. We validate both variants using medical instrument segmentation as target task.
Results
The proposed approach can be used to radically reduce the manual annotation effort involved in training CNNs. Compared to the baseline approach of generating annotated data from scratch, our method decreases exploratively the number of labeled images by up to 75% without sacrificing performance. Our method also outperforms alternative methods for CNN pre-training, such as pre-training on publicly available non-medical (COCO) or medical data (MICCAI EndoVis2017 challenge) using the target task (in this instance: segmentation).
Conclusion
As it makes efficient use of available (non-)public and (un-)labeled data, the approach has the potential to become a valuable tool for CNN (pre-)training.






Similar content being viewed by others
References
Agrawal P, Carreira J, Malik J (2015) Learning to see by moving. In: Proceedings of the IEEE international conference on computer vision
Baur C, Albarqouni S, Navab N (2017) Semi-supervised deep learning for fully convolutional networks. In: International conference on medical image computing and computer-assisted intervention, Springer
Bittel S, Roethlingshoefer V, Kenngott H, Wagner M et al (2017) How to create the largest in-vivo endoscopic dataset
Bodenstedt S, Wagner M, Katić D, Mietkowski P et al (2017) Unsupervised temporal context learning using convolutional neural networks for laparoscopic workflow analysis. arXiv:1702.03684 [cs]
García-Peraza-Herrera L.C, Li W, Gruijthuijsen C, Devreker A et al (2016) Real-time segmentation of non-rigid surgical tools based on deep learning and tracking. In: Computer-assisted and robotic endoscopy, lecture notes in computer science. Springer, Cham. https://doi.org/10.1007/978-3-319-54057-3_8
Garcia-Peraza-Herrera LC, Li W, Fidon L, Gruijthuijsen C et al (2017) Toolnet: holistically-nested real-time segmentation of robotic surgical tools. In: Proceedings of the 2017 IEEE/RSJ international conference on intelligent robots and systems
Goodfellow I, Bengio Y, Courville A (2016) Deep learning. MIT Press, Cambridge
Goodfellow I, Pouget-Abadie J, Mirza M, Xu B et al (2014) Generative adversarial nets. ArXiv: 1406.2661
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition
Heim E, Seitel A, Isensee F, Andrulis J, Stock C, Ross T, Maier-Hein L (2017) Clickstream analysis for crowd-based object segmentation with confidence. IEEE Trans Pattern Anal Mach Intell
Isola P, Zhu JY, Zhou T, Efros AA (2016) Image-to-image translation with conditional adversarial networks. arXiv preprint arXiv:1611.07004
Kamnitsas K, Baumgartner C, Ledig C, Newcombe V et al (2017) Unsupervised domain adaptation in brain lesion segmentation with adversarial networks. In: International conference on information processing in medical imaging, Springer
Larsen ABL, Sønderby SK, Larochelle H, Winther O (2016) Autoencoding beyond pixels using a learned similarity metric. In: International conference on machine learning
Larsson G, Maire M, Shakhnarovich G (2017) Colorization as a proxy task for visual understanding. arXiv:1703.04044
Lin TY, Maire M, Belongie S, Hays J et al (2014) Microsoft COCO: common objects in context. In: European conference on computer vision, Springer
Maier-Hein L, Mersmann S, Kondermann D, Bodenstedt S et al (2014) Can masses of non-experts train highly accurate image classifiers? In: Medical image computing and computer-assisted intervention - MICCAI 2014, lecture notes in computer science, Springer, Cham. https://doi.org/10.1007/978-3-319-10470-6_55
Maier-Hein L, Ross T, Gröhl J, Glocker B et al (2016) Crowd-algorithm collaboration for large-scale endoscopic image annotation with confidence. In: Medical image computing and computer-assisted intervention - MICCAI 2016, lecture notes in computer science, Springer, Cham
Maier-Hein L, Vedula SS, Speidel S, Navab N et al (2017) Surgical data science for next-generation interventions. Nat Biomed Eng 1(9):691
Mao X, Li Q, Xie H, Lau RY et al (2017) Least squares generative adversarial networks. In: 2017 IEEE international conference on computer vision (ICCV), IEEE
McCulloch CE, Neuhaus JM (2001) Generalized linear mixed models. Wiley Online Library, Hoboken
Noroozi M, Favaro P (2016) Unsupervised learning of visual representations by solving jigsaw puzzles. In: European conference on computer vision, Springer
Pakhomov D, Premachandran V, Allan M, Azizian M, Navab N (2017) Deep residual learning for instrument segmentation in robotic surgery. arXiv:1703.08580 [cs]
Pathak D, Krahenbuhl P, Donahue J, Darrell T, Efros AA (2016) Context encoders: feature learning by inpainting. In: Proceedings of the IEEE conference on computer vision and pattern recognition
Ravishankar H, Sudhakar P, Venkataramani R, Thiruvenkadam S et al (2016) Understanding the mechanisms of deep transfer learning for medical images. In: Deep learning and data labeling for medical applications, lecture notes in computer science, Springer, Cham
Ronneberger O, Fischer P, Brox T (2015) U-net: convolutional networks for biomedical image segmentation. In: International conference on medical image computing and computer-assisted intervention, Springer
Sønderby CK, Caballero J, Theis L, Shi W, Huszár F (2016) Amortised map inference for image super-resolution. arXiv preprint arXiv:1610.04490
Tajbakhsh N, Shin JY, Gurudu SR, Hurst RT, Kendall CB, Gotway MB, Liang J (2017) On the necessity of fine-tuned convolutional neural networks for medical imaging. In: Deep learning and convolutional neural networks for medical image computing, advances in computer vision and pattern recognition, Springer, Cham. https://doi.org/10.1007/978-3-319-42999-1_11
Twinanda AP, Shehata S, Mutter D, Marescaux J et al (2017) EndoNet: a deep architecture for recognition tasks on laparoscopic videos. IEEE Trans Med Imag 36:86–97
Zhang R, Isola P, Efros AA (2016) Colorful image colorization. In: European conference on computer vision, Springer
Zhang R, Isola P, Efros AA (2017) Split-brain autoencoders: unsupervised learning by cross-channel prediction. In: 2017 IEEE conference on computer vision and pattern recognition (CVPR)
Zhou Z, Shin J, Zhang L, Gurudu S et al (2017) Fine-tuning convolutional neural networks for biomedical image analysis: actively and incrementally. In: IEEE conference on computer vision and pattern recognition, Hawaii
Acknowledgements
We acknowledge the support of the European Research Council (ERC-2015-StG-37960). This work was support by Intuitive Surgical who providing us with the raw video data, from which the Medical Image Computing and Computer Assisted Intervention conference 2017 robotic challange data were extracted. We further acknowledge the support of the Federal Ministry of Economics and Energy (BMWi) and the German Aerospace Center (DLR) within the OP 4.1 projekt. Finally, we would like to thank Simon Kohl inspiring us to this paper.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
For this type of study formal consent is not required.
Informed consent
This article contains patient data from publically available datasets.
Rights and permissions
About this article
Cite this article
Ross, T., Zimmerer, D., Vemuri, A. et al. Exploiting the potential of unlabeled endoscopic video data with self-supervised learning. Int J CARS 13, 925–933 (2018). https://doi.org/10.1007/s11548-018-1772-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-018-1772-0