Abstract
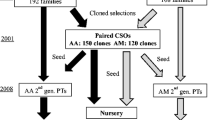
Acacia hybrids offer a great potential for paper industry in Southeast Asia due to their fast growth and ability to grow on abandoned or marginal lands. Breeding Acacia hybrids with desirable traits can be achieved through marker assisted selection (MAS) breeding. To develop a MAS program requires development of linkage maps and QTL analysis. Two mapping populations were developed through interspecific hybridization for linkage mapping and QTL analysis. All seeds per pod were cultured initially to improve hybrid yield as quality and density of linkage mapping is affected by the size of the mapping population. Progenies from two mapping populations were field planted for phenotypic and genotypic evaluation at three locations in Malaysia, (1) Forest Research Institute Malaysia field station at Segamat, Johor, (2) Borneo Tree Seeds and Seedlings Supplies Sdn, Bhd. (BTS) field trial site at Bintulu, Sarawak, and (3) Asiaprima RCF field trial site at Lancang, Pahang. During field planting, mislabeling was reported at Segamat, Johor, and a similar problem was suspected for Bintulu, Sarawak. Early screening with two isozymes effectively selected hybrid progenies, and these hybrids were subsequently further confirmed by using species-specific SNPs. During field planting, clonal mislabeling was reported and later confirmed by using a small set of STMS markers. A large set of SNPs were also used to screen all ramets in both populations. A total of 65.36% mislabeled ramets were encountered in the wood density population and 60.34% in the fibre length mapping population. No interpopulation pollen contamination was detected because all ramets found their match within the same population in question. However, mislabeling was detected among ramets of the same population. Mislabeled individuals were identified and grouped as they originated from 93 pods for wood density and 53 pods for fibre length mapping populations. On average 2 meiotically unique seeds per pod (179 seeds/93 pods) for wood density and 3 meiotically unique seeds per pod (174 seeds/53 pods) for fibre length mapping population were found. A single step statistical method was used to evaluate the most informative set of SNPs that could subsequently be used for routine checks for mislabeling in multi-location field trials and for labelling superior clones to protect breeder’s rights. A preliminary set of SNPs with a high degree of informativeness was selected for the mislabeling analysis in conjunction with an assignment test. Two subsets were successfully identified, i.e., 51 SNPs for wood density and 64 SNPs for fibre length mapping populations to identify all mislabeled ramets which had been previously identified. Mislabeling seems to be a common problem due to the complexity involved in the production of mapping populations. Therefore, checking for mislabeling is imperative for breeding activities and for analyses such as linkage mapping in which a correlation between genotypic and phenotypic data is determined.
Similar content being viewed by others
References
Adams WT (1983) Application of isozymes in tree breeding. In: Tanksley SD, Orton TJ (eds) Isozymes in plant genetics and breeding, part A. Elsevier, Amsterdam, pp 381–400
Anonymous (2009) Prepare for the deluge. Nat Biotechnol 26:1099
Asif MJ, Ariffin MAT, Yit HM, Wong M, Abdullah MZ, Muhammad N, Ratnam W (2015) Utilization of STMS markers to verify admixture in clonal progenies of Acacia mapping populations and relabeling using assignment tests. J For Sci 61(5):200–209
Asif JM, Charles HC, Wickneswari R (2016) Cross-specific amplification of microsatellite DNA markers in Shorea platyclados. J For Res 27(1):27–32
Aziah MY, McKellar D, Fadhilah Z, Halilah AK, Haliza I (1999) Establishing protocol for commercial micropropagation of Acacia mangium × Acacia auriculiformis. J Trop For Sci 11(1):148–156
Beacham TD, Spilsted B, Le KD, Wetklo M (2008) Population structure and stock identification of chum salmon Oncorhynchus keta from British Columbia determined with microsatellite DNA variation. Can J Zool 86:1002–1014
Bovo D, Rugge M, Shiao YH (1999) Origin of spurious multiple bands in the amplification of microsatellite sequences. Mol Pathol 52:50–51
Bowcock AM, Ruiz-Linares A, Romfohrde J, Minch E, Kidd JR, Cavalli-Sforza LL (1994) High resolution of human evolutionary trees with polymorphic microsatellites. Nature 368:455–457. doi:10.1038/368455a0
Butcher PA, Moran GF (2000) Genetic linkage mapping in Acacia mangium. 2. Development of an integrated map from two outbred pedigrees using RFLP and microsatellite loci. Theor Appl Genet 101:594–605
Butcher PA, Moran GF, DeCroocq S (1998a) Use of molecular markers in Cacao clone identification. Crop Sci 46:2084–2092
Butcher PA, Moran GF, Perkins HD (1998b) RFLP diversity in the nuclear genome of Acacia mangium. Heredity 81:205–213
Butcher PA, Glaubitz JC, Moran GF (1999) Applications for microsatellite markers in the domestication and conservation of forest trees. For Genet Resour Inf 27:34–42
Butcher PA, Decroocq S, Gray Y, Moran GF (2000a) Development, inheritance and cross species amplification of microsatellite markers from Acacia mangium. Theor Appl Genet 101:1282–1290
Butcher PA, Moran GF, Bell R (2000b) Genetic linkage mapping in Acacia mangium 1. Evaluation of restriction endonucleases, inheritance of RFLP loci and their conservation across species. Theor Appl Genet 100:576–583
Buttrose MS, Grant WJR, Sedgley M (1981) Floral development in Acacia pycnantha benth. Aust J Bot 29:385–395
Cabezas JA, Ibáñez J, Diego L, Dolores V, Gema B, Virginia R, Iván C, Angelica MJ, Juan C, Leonor R, Mark RT, José MM (2011) A 48 SNP set for grapevine cultivar identification. BMC Plant Biol 11:153
Chen H, Morrell PL, Vanessa ETMA, de la Marlene C, Michael TC (2009) Tracing the geographic origins of major Avocado cultivars. J Hered 100(1):56–65
Efron B (1983) Estimating the error rate of a prediction rule: improvement on cross-validation. J Am Stat Assoc 78:316–330
Fernando P, Vidya TNC, Melnick DJ (2001) Isolation and characterisation of tri- and tetranucleotide microsatellite loci in the Asian elephant (Elephas maximus). Mol Ecol Notes 1:232–233
Glover KA, Hansen MM, Lien S, Als TD, Hoyheim B, Skaala O (2010) A comparison of SNP and STR loci for delineating population structure and performing individual genetic assignment. BMC Genet 11:2
Haofeng C, Peter L, Morrell V, Ashworth ETM, Marlene DLC, Clegg MT (2009) Tracing the geographic origins of major Avocado cultivars. J Hered 100(1):56–65
Hardiyanto EB (2014) Challenges for Acacia breeders. “Sustaining the future of Acacia plantation forestry”. In: Hardiyanto EB (ed) Sustaining the future of Acacia plantation forestry held in Hue Vietnam, 18–21 March
Harju A, Muona O (1989) Background pollenisation in Pinus sylvestris seed orchards. Scand J For Res 4:513–520
Hayden MJ, Stephenson P, Logojan AM, Khatkar D, Rogers C, Koebner RMD, Snape JW, Sharp PJ (2004) A new approach to extending the wheat marker pool by anchored PCR amplification of compound SSRs. Theor Appl Genet 108:733–742
Hyten DL, Cannon SB, Song Q, Weeks N, Fickus EW, Shoemaker RC et al (2010) High-throughput SNP discovery through deep resequencing of a reduced representation library to anchor and orient scaffolds in the soybean whole genome sequence. BMC Genom 11(1):38
Jahan MS, Ahsan L, Noori A, Quaiyyum MA (2008) Process for the production of dissolving pulp from Trema orientalis (NALITA) by prehydrolysis kraft and soda-ethylenediamine (EDA) process. BioResources 3(3):816–828
Jones CJ, Edwards KJ, Castaglione S, Winfield MO, Sala F, van de Wiel C, Bredemeijer G, Vosman B, Matthes M, Daly A, Brettschneider R, Bettini P, Buiatti M, Maestri E, Malcevschi A, Marmiroli N, Aert R, Volckaert G, Rueda J, Linacero R, Vazquez A, Karp A (1997) Reproducibility testing of RAPD, AFLP and SSR markers in plants by a network of European laboratories. Mol Breed 3(5):381–390
Jones ME, Shepherd M, Henry R, Delves A (2008) Pollen flow in Eucalyptus grandis determined by paternity analysis using microsatellite markers. Tree Genet Genomes 4:37–47
Kalinowski ST (2004) Genetic polymorphism and mixed stock fisheries analysis. Can J Fish Aquat Sci 61:1075–1082
Kalinowski ST, Taper ML, Marshall TC (2007) Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol Ecol 16:1066–1099
Kawauchi H, Goto S (1999) Monitoring clonal management in nematode-resistant Japanese black pine seed orchard in Kagoshima prefecture. J Jpn For Soc 81:338–340
Keenan K, McGinnity P, Cross TF, Crozier WW, Prodöhl PA (2013) diveRsity: an R package for the estimation of population genetics parameters and their associated errors. Methods Ecol Evol 4(8):782–788
Kenrick J, Knox RB (1979) Pollen development and cytochemistry in some Australian species of Acacia. Aust J Bot 27:413–427
Kijkar S (1992) Handbook: vegetative propagation of Acacia mangium × Acacia auriculiformis. ASEAN—Canada. Forest Tree Seed Centre Project, Muak-Lek, Saraburi, Thailand
Kojima M, Yamamoto H, Okumura K, Ojio Y, Yoshida M, Okuyama T, Ona T, Matsune K, Nakamura K, Ide Y, Marsoem N, Sahri MH, Hadi YS (2009) Effect of the lateral growth rate on wood properties in fast- growing hardwood species. J Wood Sci 55:417–424
Lao O, van Duijn K, Kersbergen P, de Knijff P, Kayser M (2006) Proportioning whole-genome single-nucleotide-polymorphism diversity for the identification of geographic population structure and genetic ancestry. Am J Hum Genet 78(4):680–690
Lia VV, Bracco M, Gottlieb AM, Poggio L, Confalonieri VA (2007) Complex mutational patterns and size homoplasy at maize microsatellite loci. Theor Appl Genet 115:981–991
Liesebach H, Schneck V, Ewald E (2010) Clonal fingerprinting in the genus Populus L. by nuclear microsatellite loci regarding differences between species and hybrids. Tree Genet Genomes 6:259–269
Liew WY (2007) Development and utilisation of EST- SSR markers for genetic linkage mapping in Acacia mangium x Acacia auriculiformis hybrid. M.Sc. thesis, Universiti Kebangsaan Malaysia
Midgley S (2014) Global use of Acacia—why we are here. In: Hardiyanto EB (ed) Sustaining the future of Acacia plantation forestry held in Hue. Vietnam, 18–21 March
Millar MA, Byrne M, Nuberg I, Sedgley M (2008) High outcrossing and random pollen dispersal in a planted stand of Acacia saligna subsp. saligna revealed by paternity analysis using microsatellites. Tree Genet Genomes 4:367–377
Moriguchi Y, Tani N, Itoo S, Kanehira F, Tanaka K, Yogomida H, Taira H, Tsumura Y (2005) Gene flow and mating system in five Cryptomeria japonica D. Don seed orchard as revealed by analysis of microsatellite markers. Tree Genet Genomes 1:174–183
Moriya S, Iwanami H, Okada K, Yamamoto T, Abe K (2010) A practical method for apple cultivar identification and parent-offspring analysis using simple sequence repeat markers. Euphytica 177(1):135–150
Newman IV (1933) Studies in the Australian acacias. II. Life history of Acacia baileyana F.V.M. Part 1. Some ecological and vegetative features, spore production and chromosome number. J Linn Soc Bot 49:145–171
Ng CH, Koh S, Lee S, Ng K, Mark A, Norwati M, Wickneswari R (2005) Isolation of 15 polymorphic microsatellite loci in Acacia hybrid (Acacia mangium × Acacia auriculiformis). Mol Ecol Notes 5:572–575
Ng CH, Lee SL, Ng KKS, Muhammad N, Ratnam W (2009) Mating system and seed variation of Acacia hybrid (A. mangium × A. auriculiformis). J Genet 88(1):25–31
Nuray K, Kani I (2009) Genetic identification of clones and the genetic structure of seed crops in a Pinus brutia seed orchard. Turk J Agric For 34:127–134
Paetkau D, Slade R, Burden M, Estoup A (2004) Direct, real-time estimation of migration rate using assignment methods: a simulation-based exploration of accuracy and power. Mol Ecol 13:55–65
Paschou P, Ziv E, Burchard EG, Choudhry S, Rodriguez-Cintron W, Mahoney MW, Drineas P (2007) PCA-correlated SNPs for structure identification in worldwide human populations. PLoS Genet 3(9):1672–1686
Patterson N, Price AL, Reich D (2006) Population structure and eigen analysis. PLoS Genet 2(12):e190
Piry S, Alapetite A, Cornuet JM, Paetkau D, Baudouin L, Estoup A (2004) GeneClass2: a software for genetic assignment and first-generation migrant detection. J Hered 95:536–539
Potter K, Rimbawanto A, Beadle C (2006) Heart rot and root rot in tropical Acacia plantations. In: Proceedings of a workshop held in Yogyakarta, Indonesia. ACIAR Proceedings No. 124, Canberra, 7–9 Feb
R Development Core Team (2011) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. http://www.R-project.org
Rafalski A (2002) Applications of single nucleotide polymorphisms in crop genetics. Curr Opin Plant Biol 5:94–100
Rannala B, Mountain JL (1997) Detecting immigration by using multilocus genotypes. Proc Natl Acad Sci USA 94:9197–9201
Roda J-M, Rathi S (2006) Malaysia feeding China’s expanding demand for wood pulp: a diagnostic assessment of plantation development, fiber supply, and impacts on natural forests in China and in the South East Asia region. CIFOR. CIFOR, 23 p
Rosenberg NA, Li LM, Ward R, Pritchard JK (2003) Informativeness of genetic markers for inference of ancestry. Am J Hum Genet 73:1402–1422
Scheepers D, Elow M-C, Briquet M (1997) Use of RAPD patterns for clone verification provenance relationships in Norway spruce (Picea abies). Theor Appl Genet 94:480–485
Sedgley M, Harbard J, Smith RM (1992) Hybridization techniques for Acacias. ACIAR Technical Reports, No. 20
Smith CT, Seeb LW (2008) Number of alleles as a predictor of the relative assignment accuracy of STR and SNP baselines for chum salmon. Trans Am Fish Soc 137(3):751–762
Smith CT, Nelson RJ, Wood CC, Koop BF (2001) Glacial biogeography of North American Coho salmon (Oncorhynchus kisutch). Mol Ecol 10:2775–2785
Takrama J, Kun J, Meinhardt L, Mischke S, Opoku SY, Padi FK, Zhang D (2014) Verification of genetic identity of introduced cacao germplasm in Ghana using single nucleotide polymorphism (SNP) markers. Afr J Biotech 13(21):2127–2136
Tripathi SB, Mathish NV, Gurumurthi K (2006) Use of genetic markers in the management of micropropagated Eucalyptus germplasm. New For 31(3):361–372
Wheeler NC, Jech KS (1992) The use of electrophoretic markers in seed orchard research. New For 6:311–328
Wilkinson S, Wiener P, Archibald AL, Law A, Schnabel RD, McKay SD, Taylor JF, Ogden R (2011) Evaluation of approaches for identifying population informative markers from high density SNP chips. BMC Genet 12:45
Wong ML, Cannon CH, Wickneswari R (2012) Development of high-throughput SNP-based genotyping in Acacia auriculiformis x Acacia mangium hybrids using short-read transcriptome data. BMC Genom 13:726
Wright S (1951) The genetical structure of populations. Ann Eugen 15:323
Acknowledgements
The assistance of Nor Azurawati, Samsuri Toh Harun, Muhamad Azren Samek in field work; Lee Soon Leong, Kevin Ng Kit Siong, Mariam Din, Sharifah Talib, Ghazali Jaafar in Genetics Laboratory, FRIM; Normah Basir in Tissue Culture Laboratory, FRIM are all gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Additional information
Project funding: Financial support was provided by the Ministry of Science, Technology and Innovation Malaysia (IRPA 01-02-02-0015PR0003/03-02, 02-01-02-SF0403) and Universiti Kebangsaan Malaysia (UKM-AP-BPB-13-2009, GUP-2013-039).
The online version is available at http://www.springerlink.com
Corresponding editor: Yu Lei.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Javed Muhammad, A., Abdullah, M.Z., Muhammad, N. et al. Detecting mislabeling and identifying unique progeny in Acacia mapping population using SNP markers. J. For. Res. 28, 1119–1127 (2017). https://doi.org/10.1007/s11676-017-0405-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11676-017-0405-8