Abstract
Microbial cell surface display technology can redesign cell surfaces with functional proteins and peptides to endow cells some unique features. Foreign peptides or proteins are transported out of cells and immobilized on cell surface by fusing with anchoring proteins, which is an effective solution to avoid substance transfer limitation, enzyme purification, and enzyme instability. As the most frequently used prokaryotic and eukaryotic protein surface display system, bacterial and yeast surface display systems have been widely applied in vaccine, biocatalysis, biosensor, bioadsorption, and polypeptide library screening. In this review of bacterial and yeast surface display systems, different cell surface display mechanisms and their applications in biocatalysis as well as biosensors are described with their strengths and shortcomings. In addition to single enzyme display systems, multi-enzyme co-display systems are presented here. Finally, future developments based on our and other previous reports are discussed.
Similar content being viewed by others
Introduction
Microbial surface display technology refers to the use of gene engineering means to realize presentation of foreign peptides or proteins on the surfaces of microorganisms in the forms of fusion proteins, which can maintain their relatively independent spatial structure and biological activity [1]. As the first microbial surface display system, the phage display system was established in the mid-80s of the twentieth century, where peptides and small proteins were displayed on the phage surface by fusing with the capsid protein of filamentous phage [2]. The phage display system has many remarkable advantages, such as high-throughput biopanning, screening of mimic epitopes, simple preparation process, and so on. In recent years, this technology has had a far-reaching influence on protein molecule mutual recognition, the development of new vaccines and tumor treatments [3]. With the development of surface display technology, surface display system has been expanded from phage display system to other microbial cell display systems [4]. Currently, bacterial and yeast surface display system is the most frequently used prokaryotic and eukaryotic protein surface display system, respectively, due to unique features, including diversity of carriers, high yields of recombinant proteins, and amenability to gene engineering [5].
A cell surface display system contains three factors, namely host, carrier, and passenger. The host cells serve as the matrix to bind fusion proteins of foreign protein and an anchoring motif. The carriers are some outer membrane protein and appendant of cell surface, whose signal peptides can facilitate the transverse of passengers from intracellular to surface. The passengers are the target foreign protein, which can be displayed on the surface of cell through this technology. Microbial cell surface display technology involves the process of membrane transportation, which is closely related to the mechanism of protein secretion. There are various protein display mechanisms in different cell types (Fig. 1). Therefore, it is indispensable to coordinate with the three essential elements to establish a successful surface display system.
Some typical carriers used in Gram-negative bacterial (a), Gram-positive bacterial (b), and yeast (c) cell surface display systems. Red stars represent passengers. OM refers to outer membrane. IM refers to inner membrane. OMP refers to outer membrane protein. Lpp-OmpA chimera comprises lipoprotein and outer membrane protein. INP refers to ice nucleation protein. SpA protein refers to Staphylococcus aureus protein A. Surface layers (S-layers) are outside of the cell structure of many Archaea and Bacteria. α- and a-agglutinin can mediate the mating between yeast cells. Flocculin Flo1p is flocculating protein of Saccharomyces cerevisiae (Ref. [6,7,8])
Compared with free enzyme, cell surface-displayed enzyme has several advantages. Firstly, for the biotransformation of multi-step reaction, whole-cell catalysis can improve the efficiency of biocatalysis by coupling the cofactor and other enzymes in the cell. Secondly, the preparation of enzyme is simply achieved by culturing microbial cells, thus greatly simplifying the preparation process of the protein and reducing the production cost. Thirdly, the stability of cell surface-displayed enzyme is improved, and the whole-cell catalyst could be reusable without compromising its activity [1].
Until now, microbial surface display technology has been widely applied in all aspects of the biotechnology, such as live vaccine development, peptide library construction and screening, bioremediation of heavy metal pollution, whole-cell catalysis, and biosensors [4, 9].
Up to now, there have been several reviews related to recent advances on different surface display systems developed for phage, bacteria, and yeast [1, 10], as well as their applications in screening of protein libraries [6], vaccines [11], and medical and environmental aspects [12]. In this review, we will focus on bacterial and yeast surface display systems, as well as their applications on whole-cell biocatalysis and biosensors. Especially, constructions and performances of multi-enzyme co-display systems will be discussed.
Gram-Negative Bacterial Cell Surface Display
The cell envelope structure of Gram-negative bacteria is complex, composed of cytoplasmic membrane, periplasm, and outer membrane. In order to display on the outer membrane of cells, the foreign protein fused with the anchoring motif should pass through the cytoplasmic membrane and periplasm space. Various display systems have been developed successfully, exhibiting different presentation profiles with diverse targeting and anchoring mechanisms of carrier proteins. The Gram-negative bacterium Escherichia coli is the most frequently used host in view of its mature genetic manipulation system and high yields of biomass. Representative display systems for Gram-negative bacterial surface display systems are shown in Table 1.
Outer Membrane Protein Display System
Outer membrane proteins (OMPs) span the outer membrane through antiparallel β-sheets which form a certain 3-D structure like a barrel, such as LamB (maltoporin), OmpA (out membrane protein A), and PhoE (phosphate-inducible porin) [39, 40]. The external loops can be inserted into foreign peptide fragments. Therefore, heterologous proteins can be fused with these loops for cell surface display. Most OMP mediate display systems are limited to small foreign protein display without the disruption of membrane integrity. Fortunately, a few of systems have been developed recently to facilitate stable display of large passengers. For example, amylase (77 KDa) could be localized on the surface of E. coli using protein PgsA from Bacillus subtilis as an anchoring motif [15].
Lipoprotein Display System
Lipoprotein with the lipid-modified cysteine of N-terminus was located at the periplasmic space. TraT has been used to display foreign peptides at two fusion sites, including the middle and the C-terminus of TraT [12, 41]. The Lpp-OmpA chimera comprises the signal peptide and nine N-terminal residues of lipoprotein, as well as the residues 49-159 of OmpA [42]. The hybrid display system has been used to display several passengers, such as esterases, organophosphate hydrolases (OPH), and β-lactamase [43]. However, cell viability and enzyme activity could be influenced [44].
Ice Nucleation Protein Display System
Ice nucleation protein (INP) is an outer membrane protein in ice nucleation active bacterium, which can accelerate the formation of ice crystals in supercooled water [45]. It is involved with the genus Pseudomonas [46, 47], Erwinia [48], and Xanthomonas [49]. INP consists of three distinct domains, namely, N-terminal, C-terminal, and central repeating domain. The amino acids of the N-terminal domain are relatively hydrophobic and link the protein to the outer membrane through a glycosylphosphatidylinositol anchor, which is endowed with a particular desirable ability to display proteins (Fig. 2). Compared with other anchoring units, INP has several distinct characteristics due to its special structural and functional features: (i) It has been proved that effective bacterial surface display of foreign proteins can be achieved using INP as an anchoring motif with the full-length or truncated sequence, indicating that C-terminal and central repeating domains are not necessary for the anchoring function. Thus, surface display of passengers with large size could be realized when only N-terminal domain was served as the anchoring motif. (ii) Foreign proteins can be stably expressed in the host without degradation by intracellular or extracellular proteases. In contrast with Lpp-OmpA chimera, the cell viability will not be disrupted when INP is displayed on the surfaces of bacteria [50]. (iii) INP has secreted, guided, and anchoring functions, which could make fusion proteins self-anchored on cell surface without requiring other specific accessory protein.
Cell surface display systems using ice nucleation protein (INP) as carrier. a Display of passenger with the internal repeating domain, N domain, and C domain of INP. b Display of passenger with C domain and N domain of INP. c Display of passenger with only N domain of INP (Ref. [6])
In recently years, INP has been widely used for surface display of heterologous proteins, even large proteins, such as Chitinase 92 (90 kDa) and cytochrome P450 (119 kDa) [51], or the combination of two proteins, such as organophosphate hydrolases (OPH) and methyl parathion (MPH) [52,53,54]. In the light of these results, the successful construction of INP-mediated bacterial surface display system and its application in biocatalysis and biosensor has been implemented in our laboratory. In detail, a variety of oxidoreductases and hydrolases were displayed on the surface of E. coli using N-terminal of INP as the anchoring motif. The excellent activity and stability of prepared whole-cell biocatalysts well exhibited the remarkable performance of INP display system. Novel biosensors were constructed by using the above whole-cell biocatalysts as detection elements, realizing detection quantification of the sugar and pesticides with high sensitivity and specificity [20, 24, 26, 55, 56]. However, this technique has some limitations. For example, for passenger proteins that contain disulfide bonds in the molecule, carrier proteins can interfere with the correct folding of the passenger protein, thereby reducing the activity of the passenger.
Autotransporter Protein Display System
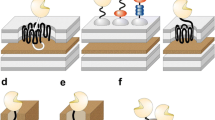
Autotransporter proteins (ATs) are related to virulence of Gram-negative bacteria, and their main functions are to adhesion and protein hydrolysis (Table 2). ATs were firstly discovered in Neisseria sp., namely IgA protease [65]. All ATs have similar structural features. The N-terminal has a signal peptide that interacts with the Sec translocon. The central is the passenger region that carries the physiological translocation function. The C-terminal is called translocation unit (TU) [66]. Most of the signal peptide is composed of three parts, charged N region, hydrophobic H region and C region, and a signal peptidase recognition site. Sec translocon is able to recognize the N and H regions of the signal peptide to direct the polypeptide chain through the inner membrane into the periplasmic space, and then signal peptide is hydrolyzed and removed under the catalysis of the signal peptidase. The size and function of the passenger region are very different, and the passenger area affects and determines the biological function of ATs. More than 97% of ATs are structurally right-handed beta-helices, which will correctly fold on cell surface after crossing the membrane, while the passenger region is away from the bacterial surface. The function of TU is to transport the passenger from the periplasmic space to the bacterial surface. At present, the 3-D structures of various TU have been reported, such as extracellular serine protease (EspP), adhesion involved in diffuse adherence I (AIDA-I) and EstA from P. aeruginosa, and so on. TU structure is basically similar, consisting of a β-barrel structure and an extended alpha-helix. The β-barrel structure consists of 12 consecutive β-sheets, and the α-helix is located in the inner cavity of the β-barrel structure. The adjacent two beta-sheets are linked together by a β-Turn or a cell exposed to an extracellular loop. Extracellular loop, β-turn, and α-helix can prevent the loss of intracellular material through the channel, but also to prevent the extracellular harmful substances (such as antibiotics, etc.) through the channel into the cell.
All the ATs use V-type secretory pathway. The secretory process consisted of four steps, including inner membrane transport, periplasmic transport, outer membrane transport, and passenger region modification [66]. The specific cell surface display mechanism using ATs as carrier was shown in Fig. 3. Many studies showed that AT was an ideal tool to display the exogenous proteins due to the fact that this system does not require the participation of other proteins and can transport larger size proteins as well as maintain their antigenic and biological functions [67]. For example, the outer membrane esterase EstA proved to successfully direct lipase and its specific chaperone to the microbial cell surface [36]. Besides, AT display system is highly efficient. The study has shown that ATs can exhibit more than 250,000 passenger protein on a single bacterial surface [34]. Coupling with flow cytometric analysis, the AT-mediated surface display system can be used to screen large libraries of variants generated by directed evolution. Although some models have been put forwarded to explain transport process of passenger region from periplasm to outer membrane, including the threading model, the multimeric model, the hairpin model, and so on, none of the above models can explain comprehensively the whole process, and part of the assumption is also the lack of experimental data support.
The specific cell surface display mechanism using ATs as carrier. Sec translocon can recognize the signal peptide to direct the polypeptide chain through the inner membrane into the periplasmic space, and then signal peptide is hydrolyzed and removed under the catalysis of the signal peptidase. TU is folded into a β-barrel structure, which is then inserted into the extracellular membrane to form a channel through which the passenger is transported to the bacterial surface (Ref. [66])
Gram-Positive Bacterial Cell Surface Display
Although Gram-positive bacteria have thicker and tougher outer cell wall than Gram-negative bacteria, proteins are presented on the surfaces of Gram-positive bacteria only through a single membrane layer, avoiding negative influences on the structure and activity of protein during the traverse through the periplasmic space and the integration on the outer membrane. However, low transformation efficiency will hinder the construction of the peptide library with large capacity.
Staphylococcus aureus Protein A Display System
The C-terminal sequence of SpA can be anchored to the bacterial surface through the combination of a hydrophobic region with the peptidoglycan of cell wall [68]. The N-terminal is allowed to be inserted or replaced by foreign protein to achieve surface display. SpA consists of a signal peptide, an IgG-binding domain, and a C-terminal surface-binding domain. The C-terminal region consists of a charged repeating region containing LPXTG motif, a hydrophobic region, and a short charged tail RREL [69]. Staphylococcal surface display system utilizes the C-terminal anchoring region of SpA, replacing the IgG-binding domain with an exogenous protein, which is expressed as a fusion protein and displayed on the cell surface [70].
Surface Layer Display System
Surface Layers (S-layers) are outside of the cell structure of many Archaea and Bacteria, which is composed of a single protein or glycoprotein subunit and arranged regularly in the cell membrane or cell wall [71]. Amino acid sequence analysis showed that the S-layers of Lactobacillus acidophilus contain two conserved regions, including N-terminal signal sequence and C-terminal region, which can make foreign protein anchor on the surface of cell [72]. S-layers have been widely used in the construction of highly efficient secretory expression vector owing to their high level of transcription, expression, and secretion. Therefore, lactic acid bacterium surface display system was established using S-layers of Lactobacillus as carrier to display foreign antigens outside the cell surface, in order to avoid degradation by cytoplasmic protease [73].
Bacillus subtilis Spore Surface Display System
Bacillus subtilis spore surface display system is an important Gram-positive bacterial cell surface display system with spore coat protein as carrier, including CotB, CotC, CotG, OxdD, and other capsid proteins (Table 3). Studies showed that different carriers would lead to different display efficiencies [77]. The fusion of certain foreign proteins with the anchoring protein CotB does not affect the surface display [83]. But for some foreign proteins, the N-terminal fusion may be more reasonable. Isticato et al. displayed foreign protein using N-terminal and C-terminal of CotC protein. The result showed N-terminal fusion method could detect CotC monomer and dimer, but C-terminal fusion method could only detect a small amount of CotC monomer. Comparing with C-terminal fusion method, the display efficiency of N-terminal fusion method increased five times. The reason for this phenomenon may be that the C-terminal of CotC needs to interact with other CotC or capsid proteins during spore formation [84].
Yeast Cell Surface Display
Bacterial surface display system is incapable of expressing complex eukaryotic proteins, which need post-translational modifications to exhibit activity, including glycosylation and disulfide isomerization. Instead, as a eukaryotic system, Saccharomyces cerevisiae had been successfully employed to express and display dozens of complex proteins in the past decades, which could offer an easy handling procedure, stable activity of expressed enzymes, and feasibility to construct large protein libraries. In general, the target proteins were expressed in a fusion form with cell wall proteins, which were secreted out of the cell and then interact with the cell wall via a covalent or non-covalent linkage.
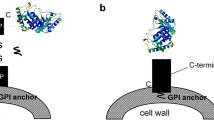
S. cerevisiae carrier protein can be divided into C-terminal anchored carrier protein and N-terminal anchored carrier protein according to its binding to cell wall (Table 4). C-terminal anchored carrier protein is mainly glycosylphosphatidylinositol (GPI) protein, such as α-agglutinin [95], Cwp1p [96], Cwp2p, and C-terminal of Flo1p [90], in which the N-terminal fragment was fused to passenger protein and C-terminal GPI anchor could covalently link to the cell wall through β-1,6-glucan bridge (Fig. 4a, b). N-terminal anchored carrier protein has three types, including a-agglutinin [97], N-terminal of Flo1p [98], and Pir protein [99]. The a-agglutinin is composed of two domains, namely Aga1 and Aga2. Aga1 could covalently link to the cell wall through GPI anchor. Aga2 binds to Aga1 by the disulfide bond, and the passenger could be display on cell surface by fusing with the C-terminal of Aga2 (Fig. 4c). But the display efficiency depends on the expression level of Aga1. The N-terminal of Flo1p is a flocculating domain that recognizes and is non-covalently bound to the mannan component in the cell wall, causing the cells to accumulate (Fig. 4d). Pir alternatively interacts with β-1,3-glucan in the cell wall via a ester bond. However, there is a lack of literature to show the display efficiency of these anchors which may vary dependent on the target proteins.
Yeast surface display systems, including α-agglutinin (a), C-terminal of Flo1p (b), a-agglutinin (c), and N-terminal of Flo1p (d). α-agglutinin and C-terminal of Flo1p are C-terminal anchored carrier proteins, in which the N-terminal fragments were fused to passenger proteins and C-terminal GPI anchor could covalently link to the cell wall through β-1,6-glucan bridge. a-agglutinin and N-terminal of Flo1p are N-terminal anchored carrier proteins, in which the C-terminal fragments were fused to passenger proteins (Ref. [8])
Applications of Microbial Surface Display in Biocatalysis
Biocatalyst generally refers to free or immobilized enzymes and living cells, which could be used to catalyze certain reaction with high efficiency under normal temperature and pressure. Microbial surface display system facilitates the development of new biocatalyst. Various enzymes have been presented on the surfaces of cells with favorable catalytic activity and stability.
Construction of Biocatalyst for Detoxification of Organophosphates
OPH was firstly displayed on the surface of E. coli JM105 by Lpp-OmpA display system, and the results showed that catalytic efficiency of whole cell was seven times higher than that of free enzyme towards parathion and paraoxon [100]. Unfortunately, the growth of cells could be inhibited grossly. Then, researchers targeted OPH on the surface of E. coli and Pseudomonas putida using truncated INP as anchoring unit in order to overcome this drawback. The results showed that enzyme activity of OPH displayed on the surface of P. putida was ten times higher than that on E. coli cell surface, probably because of the similar secretary channel between OPH original strain Flavobaeterium sp. ATCC27551 and P. diminuta MG [44, 101]. S. cerevisiae also has been used as the host for surface display of OPH in consideration of practical industrial application, which exhibited about sixfold higher activity than INP display system [102]. In our previous work [24], OPH mutant S5 with improved enzyme activity was successfully displayed on the surface of E. coli using INP as anchoring motif and its level of enzyme activity (12.44 U/mg cells) was greatly higher than those of other reports (< 1 U/mg cells). The whole-cell catalyst was a multifunctional bacterium that could be used for degradation and detection of organophosphate pesticides. In subsequent work, at the aim of enhancing the activity and stability of OPH-fused whole-cell biocatalysts, the novel cell-inorganic hybrid materials were explored by the combination of microbial cell surface display technology and biomineralization (Fig. 5). Cell-displayed OPH showed the allosteric effect from “inactive” form to “active” form and the “active” form would be “fixed” when cell-displayed OPH was embedded into cobalt phosphate crystals. Therefore, the activity of mineralized OPH-fused cells was significantly enhanced to three times higher than that of original OPH-fused cells. As a bonus, the stability of the novel hybrid catalysts was also significantly improved. Further, the as-synthesized bio-inorganic hybrid catalysts could be applied on high-efficiency degradation and sensitive detection of paraoxon. To our knowledge, this work is the first example on the cell surface-displayed enzyme-inorganic hybrid biocatalysts and would provide a model method to develop a wide range of whole-cell biocatalysts for the diverse applications on industrial catalysis, environmental governance, and biosensing [103].
The bio-inorganic hybrid whole-cell catalysts are fabricated for sensitive biosensing of organophosphorus pesticides by combination of biomineralization and microbial cell surface display technology. Due to the allosteric effect of organophosphorus hydrolase (OPH) embedded into inorganic crystal (Co3(PO4)2∙8H2O) from “inactive” form to “active” form, the biomineralized OPH-fused cells show the enhanced catalytic activity
Cell Surface Display of Lipase for Whole-Cell Biocatalysis
Since 1994, lipase has been displayed on the surfaces of cells based on various display systems. The common host and carrier were E coli and OMPs, including OmpC, FadL, and OprF [104]. Activity of whole cell reached 2800 U/mg dry cells when lipase from Bacillus sp. strain TG43 presented on the surface of bacterium using OprF as carrier [105]. Lipase yeast display focused on α-agglutinin and Flo1p display system. Rhizopus oryzae lipase was first presented on the surface of yeast using N-terminal of Flo1p, which showed 14-fold higher activity (61.3 U/g dry cell weight) than α-agglutinin (4.1 U/g dry cell weight) [91]. Becker has found that the display efficiency of lipase and cell survival rate would decrease with the increasing molecular weight of target protein [106]. Compared with free lipase, the cell surface-displayed lipase showed superior stability towards high temperature and organic solvent [107]. Recently, new anchor proteins were developed, which included Flo9 and Pir1 from Pichia pastoris, and were able to display lipase B on yeast surface for hydrolysis of tributyrin. The constructed whole-cell biocatalyst showed conservation of ~ 40% of its original activity after 3 h incubation at 45 °C, while the free enzyme lost 60% of its original activity after 10 min at 45 °C [108]. Until now, display of lipase has been mainly applied to enantioselective resolution of racemic compounds. In addition, biodiesel could be catalyzed using cell surface-displayed lipase as whole cell biocatalyst [91, 109].
Construction of Multi-Enzyme Co-displayed Systems for Bioethanol Production
Compared to some yeast species such as P. pastoris and Hansenula polymorpha, S. cerevisiae has superior fermentation characteristics, especially high cell density culture on an economical carbon source. These features permit scientists’ attempt to display enzymes involved in the decomposition of complex polysaccharides, such as cellulose and starch. S. cerevisiae could also be used to display phytase, and average ethanol fermentation rate of the resulting whole-cell biocatalyst improved 18% compared to the control strain using grain as raw material [110].
Originally, the major components of cellulosome, including β-glucosidases, endoglucanases cellobiohydrolases, and carboxymethyl cellulase, were displayed on yeast surface independently [111, 112]. To effectively hydrolyze cellulose, these enzymes were co-displayed on yeast surface to form a multi-enzyme catalytic system. The robust yeast display (YSD) system using a-agglutinin receptor as anchoring motif helped to achieve this purpose [113]. Actually, multiple enzyme encoding genes were ligated to Aga2 gene in one vector and then these enzymes were secreted out of cells to bind on Aga1 resulting in enzyme random co-display [114, 115]. To further control the ratio and exact position on cell wall of the displayed enzymes, the high-affinity interactions between cohesins and dockerins, by which the cellulase components could be held together forming the structural scaffoldin of cellulosome, were employed in YSD system. In this system, a series of cohesins domains were ligated together with the controlled space and fused with Aga2 to bind on yeast cell wall. The displayed enzymes were fused with different dockerins and secreted out of the cell, which then interacted with the cohesin domains in a specific manner [116]. This system allows a series of designed reactions on one whole-cell biocatalyst.
It should be recognized that most of the YSD systems developed for starch and cellulose hydrolysis were based on S. cerevisiae as it has the ability to produce ethanol using these carbon sources. Incomplete self-assembly of heterologous minicellulosome on the yeast cell surface was first realized in 2009, and the engineered yeast was capable of hydrolyzing phosphoric acid-swollen cellulose with several times higher ethanol level than that reached by using purified enzymes (1.2–3.5 g/L) [117, 118]. Moreover, yeast surface display of self-assembled minicellulosomes for simultaneous saccharification and fermentation of cellulose to ethanol was reported for the first time in 2010, which generated 1.8 g/L of ethanol [119]. Self-assembly of cellulosomes with two miniscaffoldins on the yeast cell surface was put forwarded in order to increase the production of ethanol based on above researches (Fig. 6). The study used cellulases from mesophilic strain in consideration of thermal compatibility between cellulose hydrolysis and yeast fermentation. The display level of cellulosome increased, and direct fermentation of microcrystalline cellulose to ethanol was realized [120]. Nevertheless, the production of ethanol is still low. The probably reason was excessive metabolic burden on the host and limitation of surface assembly when so many complex and large proteins were simultaneously expressed in cells and presented on the surface. In addition, the self-assembly of cellulosomes needs to be done under the induction of galactose, but the presence of glucose inhibits the use of galactose by yeast cells. So, the assembly process and the fermentation process must be carried out using galactose and cellulose as carbon, respectively, which would decrease cell cellulose saccharification capacity in whole fermentation process.
Cellulosomes with two miniscaffoldins self-assembly on the yeast cell surface. Cellulases displayed in this system are as follows: endoglucanase (EG), cellobiohydrolase (exoglucanase) (CBH), and β-glucosidase (BGL). Type I dockerins are as follows: DocI-1, DocI-2, and DocI-3. Type I cohesions are as follows: CohI-1, CohI-2, CohI-3. Type II cohesions are as follows: CohII-(1), CohII-(2), CohII-(repeating number) (Ref. [120])
Applications of Microbial Surface Display in Biosensors
Highly Selective and Sensitive Saccharide Biosensors Based on Oxido-Reductase Surface-Displayed Microorganism
Carbohydrate compounds are not only the main source of energy needed for the maintenance of life activities, but are also important industrial raw materials. Therefore, saccharide analysis in food, medicine, and biological processes is extremely important. Traditional detection methods of saccharides suffer from two major drawbacks, i.e., inaccuracy and complicated operation [19]. Several novel biosensors constructed by using enzyme surface-displayed whole cells and nanostructured materials established a rapid and simple method to detect saccharides with high sensitivity and selectivity in our laboratory. Generally, oxido-reductase catalyzed saccharides were displayed on the surfaces of bacteria and yeasts by fusing with anchoring motif, such as INP and a-agglutinin [25, 55]. Multi-walled carbon nanotubes (MWNTs) and oxido-reductase surface displayed cells were cast onto electrode, and concentration of saccharide was determined through measuring the redox signals originating from the above electrode [56]. The resulting biosensor exhibited good stability, wide linear range, and lower detection limit as well as certain anti-interference ability (Fig. 7) [19]. In order to further improve performance of biosensors, rational designs of cell surface-displayed enzymes for increasing stability and selectivity were carried out in our following studies [121]. For example, Nafion/bacteria-displaying glucose dehydrogenase (GDH) mutant/MWNTs composite film-modified electrode was constructed to determine the concentration of glucose [20]. The biosensor prepared with GDH mutant Q252L/E170R/V149K/G259A exhibited good operational stability. Specifically, the amperometric response drifted less than 7% over a period of 1 h, comparing with the wild-type one (10% signal drift during 25 min recording).
The construction of a sensitive xylose electrochemical biosensor based on xylose dehydrogenase (XDH) displayed cells and multi-walled carbon nanotubes modified electrode. (a) Cell surface-displayed XDH could convert xylose to D-xylonolactone, with NAD+ as its cofactor. (b) Current-time curve obtained for the Nafion/XDH-bacteria/MWNTs/GCE on the successive addition of xylose in 0.1 M PBS (pH 7.4) containing 2 mM NAD+, on which the xylose concentration labeled is the xylose concentration in the PBS solution. Applied potential, +0.5 V vs. Ag/AgCl. (c) Typical calibration graph of the biosensor for xylose. (Ref. [19])
Sensitive Electrochemical Microbial Biosensor for Detection of Toxicant
As the most widely used pesticides in the world, organophosphate (OP) insecticides residues are the major causes of food poisoning. A variety of methods have been developed to detect OP pesticides, but there is still no fast, low-cost, and portable approach. As mentioned above, OPH-displayed strain has been exhibiting good detoxification ability. Thus, the whole-cell biocatalyst could be also used to determine the concentration of OP pesticides. In our previous work, a biosensor based on cell surface-displayed OPH and ordered mesopore carbons for highly specific, sensitive, and rapid detection of p-nitrophenyl-substituted OPs compounds was developed [122]. Although, OPH and methyl parathion hydrolase (MPH)-green fluorescent protein (GFP) fusion were co-displayed on the cell surface of E. coli using the INP and Lpp-OmpA as the anchoring motifs to further expanded the analyte range in other laboratory, the low detection limit was 100-fold higher than our results [123]. Besides OPH and MPH, acetylcholinesterase (AChE) yeast expressed system could simultaneously detect organic phosphorus and carbamate pesticides with higher sensitivity [124]. However, electrochemical biosensor based on cell surface-displayed AChE was not reported so far. Thus, further study should be focused on the construction of biosensor by integrating AChE-displayed yeast and nanostructured materials, and current efforts in our laboratory are exploring this possibility. In addition to pesticides, toxic heavy metals also threaten safety and security of humans. In order to develop simple, portable, inexpensive, and rapid analytical methods for on-site detecting toxic heavy metals, phytochelatin synthase surface-displayed yeast cell was applied to cadmium biosensors. Unfortunately, the proposed method had room for improvement in terms of sensitivity and selectivity [125].
Application of Microbial Surface Display on Medical Diagnosis
Genetically engineered microbial diagnostic biosensors offer greater sensitivity and convenience compared to classical techniques. Endocrine disrupting compounds (EDCs) are some chemical compounds with similar estrogen activity but different chemical structures, resulting in difficult measurement. To address this issue, Furst et al. developed a novel biosensor based on estrogen receptor alpha (ERα) surface-displayed bacteria, which enabled detections of many detrimental compounds. With this fast and portable device, sub-ppb levels of estradiol and ppm levels of bisphenol A are detected in complex samples. In detail, the novel electrochemical sandwich assay composed of monobodies assembled gold electrode and ERα surface-displayed bacteria. Changes in the surface impedance could be detected in the presence of EDCs because cells could bind onto EDCs captured gold electrode through the recognition between surfaced-displayed ERα and EDCs. Compared to free ERα composed system, the whole cell diagnostic biosensor showed about 100-fold improved sensitivity, which was due to a substantially increased impedance response from the binding of the large cells to the gold surface. Therefore, the small sample size and simple detection system facilitate its application in the field, and this approach was adequate for measurement of other diverse families of compounds that bind to a single receptor, such as PPARγ [126]. Similar strategy was also used to detect Salmonella outer membrane protein antigen (Fig. 8). Yeast cells were designed to display both single chain variable fragment (scFv) antibodies and gold-binding peptide (GBP) on their surfaces using a-agglutinin as anchoring motif. Sensitive detection of Salmonella TM43-E10 surface antigen was realized through sandwich format where the antigen is specifically captured at the cell surface [127]. In order to improve detection sensitivity, the scFv-displayed yeast cell could be fragmented using mortar and pestle with the scFv complex remaining intact to generate nanoyeast-scFvs, which can eliminate interference of larger fragments with electrochemical-based biosensors [128]. Therefore, these studies demonstrated the potential for microbial diagnostic biosensors to serve as robust and low-cost alternatives to current protein-based diagnostic methods.
Schematic illustration of “whole-cell” display of affinity molecules. Step I: After induction, yeast cells are spun and washed and then directly affixed to metal electrodes. Step II: Sample is mixed with detection reagents and applied to the prepared electrode surface and, after incubation, the sample is washed with buffer containing the final detection substrate and redox active molecules. Step III: Detection is carried out on a laboratory or mobile potentiostat (Ref. [127])
Conclusion and Prospects
In conclusion, compared with traditional enzymes, surface-displayed enzymes have many advantages. First of all, it is easy to cell culture with high yield and low cost. The copy number of recombinant protein is generally high. Secondly, the enzyme will be inevitably inactivated during the catalytic process or in appropriate environment, whereas the bacterial surface-displayed enzyme can be regenerated through bacterial proliferation. Thirdly, compared with the intracellular enzyme, the cell surface-displayed enzyme exhibits excellent catalytic efficiency and stability. However, the application of this technology is mainly in the laboratory research stage, and the existing surface display system still has shortcomings, such as size limit of passenger and low efficiency of display. Therefore, the development of surface display systems will be the focus of surface display technology research.
As discussed above, carriers in microbial cell surface display system are proteins of outer membrane or cell wall. So, analysis of the proteome of outer membrane or cell wall will be conducive to explore new anchoring motifs. Moreover, artificial display platform could be constructed on the base of well-known carriers, such as spore coat and lipid bilayers. Up to now, the majority of passengers are displayed on surfaces of bacteria using pathogenic and non-food grade strains as host due to their mature genetic manipulation system, especially E. coli [129]. However, this kind of bacteria could hardly be employed in some practical applications, such as in situ degradation of chemical pollutants in the process of food fermentation or antigen delivery vehicle in human body. Therefore, in order to facilitate the presentation of passengers on the surfaces of food-grade, noninvasive, and nonpathogenic bacteria, versatile genetic manipulation and foreign protein expression platforms should be developed through gene engineering means.
Furthermore, the analysis of the tertiary structure of anchoring proteins and in-depth understanding of the role of each component in the transport of passenger protein will help to guide the construction of the cell surface display system and to achieve the functional expression of protein (enzyme). For example, the tertiary structure of several ATs has been analyzed, expounding the role of C-terminal of conserved β-barrel structure in transportation of N-terminal passenger region. However, there are still many unsolved questions about the secretion and folding mechanisms of ATs display system. For example, how is the β-barrel structure of ATs integrated into the outer membrane of the bacteria? The interaction between the passenger region and TU is still lacking in-depth research. Therefore, the further analysis of the tertiary structure of ATs will provide insight into the role of the linked peptide in the interaction between the passenger region and TU, helping to design more reasonable link peptide in the construction of surface display system, realizing the functional display of the proteins (enzymes).
It is worth noting that more kinds of enzymes with different catalytic activities should be displayed on cell surface for various biocatalysis and biosensor. For example, biomass-degradable enzymes could be displayed on surface of E. coli or S. cerevisiae for production of bioenergy or biochemicals. With the development of biomineralization [130,131,132,133] and nanotechnology [134, 135], the integration of cell surface display and versatile inorganic nanomaterials would not only expand scope of the analytes, but also fundamentally improve activity and stability of surface-displayed enzyme as well as sensitivity and stability of biosensor. In addition, so far, present cell surface display-based assays are confined to electrochemical biosensor and spectrophotometric assay. More kinds of analytical methods should be integrated with cell surface display.
In a word, with the development of molecular biology technology and nanotechnology, bacterial and yeast surface display technology will play more and more important role in the fields of biocatalysis and biosensor.
References
Lee, S. Y., Choi, J. H., & Xu, Z. (2003). Microbial cell-surface display. Trends in Biotechnology, 21(1), 45–52. https://doi.org/10.1016/S0167-7799(02)00006-9.
Smith, G. P. (1985). Filamentous fusion phage—novel expression vectors that display cloned antigens on the virion surface. Science, 228(4705), 1315–1317. https://doi.org/10.1126/science.4001944.
Aghebati-Maleki, L., Bakhshinejad, B., Baradaran, B., Motallebnezhad, M., Aghebati-Maleki, A., Nickho, H., Yousefi, M., & Majidi, J. (2016). Phage display as a promising approach for vaccine development. Journal of Biomedical Science, 23(1), 66. https://doi.org/10.1186/s12929-016-0285-9.
Ueda, M. (2016). Establishment of cell surface engineering and its development. Bioscience Biotechnology and Biochemistry, 80(7), 1243–1253. https://doi.org/10.1080/09168451.2016.1153953.
Charbit, A., Boulain, J. C., Ryter, A., & Hofnung, M. (1986). Probing the topology of a bacterial-membrane protein by genetic insertion of a foreign epitope—expression at the cell-surface. EMBO Journal, 5(11), 3029–3037.
Van Bloois, E., Winter, R. T., Kolmar, H., & Fraaije, M. W. (2011). Decorating microbes: surface display of proteins on Escherichia coli. Trends in Biotechnology, 29(2), 79–86. https://doi.org/10.1016/j.tibtech.2010.11.003.
Desvaux, M., Dumas, E., Chafsey, I., & Hebraud, M. (2006). Protein cell surface display in Gram-positive bacteria: from single protein to macromolecular protein structure. FEMS Microbiology Letters, 256(1), 1–15. https://doi.org/10.1111/j.1574-6968.2006.00122.x.
Sheehan, J., & Marasco, W. A. (2015). Phage and Yeast Display. Microbiology Spectrum, 3(1), AID-0028-2014. https://doi.org/10.1128/microbiolspec.AID-0028-2014.
Chen, H., Ullah, J., & Jia, J. (2017). Progress in bacillus subtilis spore surface display technology towards environment, vaccine development, and biocatalysis. Journal of Molecular Microbiology and Biotechnology, 27(3), 159–167. https://doi.org/10.1159/000475177.
Jahns, A. C., & Rehm, B. H. A. (2012). Relevant uses of surface proteins—display on self-organized biological structures. Microbial Biotechnology, 5(2), 188–202. https://doi.org/10.1111/j.1751-7915.2011.00293.x.
Georgiou, G., Stathopoulos, C., Daugherty, P. S., Nayak, A. R., Iverson, B. L., & Curtiss, R. (1997). Display of heterologous proteins on the surface of microorganisms: from the screening of combinatorial libraries to live recombinant vaccines. Nature Biotechnology, 15(1), 29–34. https://doi.org/10.1038/nbt0197-29.
Samuelson, P., Gunneriusson, E., Nygren, P. A., & Stahl, S. (2002). Display of proteins on bacteria. Journal of Biotechnology, 96(2), 129–154. https://doi.org/10.1016/S0168-1656(02)00043-3.
Wang, J. A., Michel, V., Leclerc, C., Hofnung, M., & Charbit, A. (1999). Immunogenicity of viral B-cell epitopes inserted into two surface loops of the Escherichia coli K12 LamB protein and expressed in an attenuated aroA strain of Salmonella typhimurium. Vaccine, 17(1), 1–12.
Agterberg, M., Adriaanse, H., Vanbruggen, A., Karperien, M., & Tommassen, J. (1990). Outer-membrane Phoe protein of Escherichia-Coli K-12 as an exposure vector—possibilities and limitations. Gene, 88(1), 37–45. https://doi.org/10.1016/0378-1119(90)90057-X.
Narita, J., Okano, K., Tateno, T., Tanino, T., Sewaki, T., Sung, M. H., Fukuda, H., & Kondo, A. (2006). Display of active enzymes on the cell surface of Escherichia coli using PgsA anchor protein and their application to bioconversion. Applied Microbiology and Biotechnology, 70(5), 564–572. https://doi.org/10.1007/s00253-005-0111-x.
Cho, C. M. H., Mulchandani, A., & Chen, W. (2004). Altering the substrate specificity of organophosphorus hydrolase for enhanced hydrolysis of chlorpyrifos. Applied and Environmental Microbiology, 70(8), 4681–4685. https://doi.org/10.1128/AEM.70.8.4681-4685.2004.
van Bloois, E., Winter, R. T., Janssen, D. B., & Fraaije, M. W. (2009). Export of functional Streptomyces coelicolor alditol oxidase to the periplasm or cell surface of Escherichia coli and its application in whole-cell biocatalysis. Applied Microbiology and Biotechnology, 83(4), 679–687. https://doi.org/10.1007/s00253-009-1904-0.
Wu, P. H., Giridhar, R., & Wu, W. T. (2006). Surface display of transglucosidase on Escherichia coli by using the ice nucleation protein of Xanthomonas campestris and its application in glucosylation of hydroquinone. Biotechnology and Bioengineering, 95(6), 1138–1147. https://doi.org/10.1002/bit.21076.
Li, L., Liang, B., Shi, J. G., Li, F., Mascini, M., & Liu, A. H. (2012). A selective and sensitive D-xylose electrochemical biosensor based on xylose dehydrogenase displayed on the surface of bacteria and multi-walled carbon nanotubes modified electrode. Biosensors & Bioelectronics, 33(1), 100–105. https://doi.org/10.1016/j.bios.2011.12.027.
Liang, B., Lang, Q., Tang, X., & Liu, A. (2013). Simultaneously improving stability and specificity of cell surface displayed glucose dehydrogenase mutants to construct whole-cell biocatalyst for glucose biosensor application. Bioresource Technology, 147, 492–498. https://doi.org/10.1016/j.biortech.2013.08.088.
Gao, F., Ding, H. T., Feng, Z., Liu, D. F., & Zhao, Y. H. (2014). Functional display of triphenylmethane reductase for dye removal on the surface of Escherichia coli using N-terminal domain of ice nucleation protein. Bioresource Technology, 169, 181–187. https://doi.org/10.1016/j.biortech.2014.06.093.
Vanderveer, T. L., Choi, J., Miao, D., & Walker, V. K. (2014). Expression and localization of an ice nucleating protein from a soil bacterium, Pseudomonas borealis. Cryobiology, 69(1), 110–118. https://doi.org/10.1016/j.cryobiol.2014.06.001.
Wang, W., Zhang, Z., Ni, H., Yang, X. M., Li, Q. Q., & Li, L. (2012). Decolorization of industrial synthetic dyes using engineered Pseudomonas putida cells with surface-immobilized bacterial laccase. Microbial Cell Factories, 11(1), 75. https://doi.org/10.1186/1475-2859-11-75.
Tang, X. J., Liang, B., Yi, T. Y., Manco, G., Palchetti, I., & Liu, A. H. (2014). Cell surface display of organophosphorus hydrolase for sensitive spectrophotometric detection of p-nitrophenol substituted organophosphates. Enzyme and Microbial Technology, 60, 80–80. https://doi.org/10.1016/j.enzmictec.2014.04.006.
Wang, H. W., Lang, Q. L., Li, L., Liang, B., Tang, X. J., Kong, L. R., Mascini, M., & Liu, A. H. (2013). Yeast surface displaying glucose oxidase as whole-cell biocatalyst: construction, characterization, and its electrochemical glucose sensing application. Analytical Chemistry, 85(12), 6107–6112. https://doi.org/10.1021/ac400979r.
Liang, B., Zhang, S., Lang, Q. L., Song, J. X., Han, L. H., & Liu, A. H. (2015). Amperometric L-glutamate biosensor based on bacterial cell-surface displayed glutamate dehydrogenase. Analytica Chimica Acta, 884, 83–89. https://doi.org/10.1016/j.aca.2015.05.012.
Song, J., Liang, B., Han, D., Tang, X., Lang, Q., Feng, R., Han, L., & Liu, A. (2015). Bacterial cell-surface displaying of thermo-tolerant glutamate dehydrogenase and its application in L-glutamate assay. Enzyme and Microbial Technology, 70, 72–78. https://doi.org/10.1016/j.enzmictec.2014.12.002.
Li, Q. Q., Yu, Z. N., Shao, X. H., He, J., & Li, L. (2009). Improved phosphate biosorption by bacterial surface display of phosphate-binding protein utilizing ice nucleation protein. FEMS Microbiology Letters, 299(1), 44–52. https://doi.org/10.1111/j.1574-6968.2009.01724.x.
Kang, S. M., Rhee, J. K., Kim, E. J., Han, K. H., & Oh, J. W. (2003). Bacterial cell surface display for epitope mapping of hepatitis C virus core antigen. FEMS Microbiology Letters, 226(2), 347–353. https://doi.org/10.1016/S0378-1097(03)00623-2.
Francisco, J. A., Earhart, C. F., & Georgiou, G. (1992). Transport and anchoring of Beta-lactamase to the external surface of Escherichia-Coli. Proceedings of the National Academy of Sciences of the United States of America, 89(7), 2713–2717. https://doi.org/10.1073/pnas.89.7.2713.
Chang, H. H., Sheu, S. Y., & Lo, S. C. J. (1999). Expression of foreign antigens on the surface of Escherichia coli by fusion to the outer membrane protein TraT. Journal of Biomedical Science, 6(1), 64–70.
Rizos, K., Lattemann, C. T., Bumann, D., Meyer, T. F., & Aebischer, T. (2003). Autodisplay: efficacious surface exposure of antigenic UreA fragments from Helicobacter pylori in Salmonella vaccine strains. Infection and Immunity, 71(11), 6320–6328. https://doi.org/10.1128/IAI.71.11.6320-6328.2003.
Kranen, E., Detzel, C., Weber, T., & Jose, J. (2014). Autodisplay for the co-expression of lipase and foldase on the surface of E-coli: washing with designer bugs. Microbial Cell Factories, 13(1), 19. https://doi.org/10.1186/1475-2859-13-19.
Detzel, C., Maas, R., Tubeleviciute, A., & Jose, J. (2013). Autodisplay of nitrilase from Klebsiella pneumoniae and whole-cell degradation of oxynil herbicides and related compounds. Applied Microbiology and Biotechnology, 97(11), 4887–4896. https://doi.org/10.1007/s00253-012-4401-9.
Yang, T. H., Kwon, M.-A., Song, J. K., Pan, J. G., & Rhee, J. S. (2010). Functional display of Pseudomonas and Burkholderia lipases using a translocator domain of EstA autotransporter on the cell surface of Escherichia coli. Journal of Biotechnology, 146(3), 126–129. https://doi.org/10.1016/j.jbiotec.2010.01.022.
Wilhelm, S., Rosenau, F., Becker, S., Buest, S., Hausmann, S., Kolmar, H., & Jaeger, K. E. (2007). Functional cell-surface display of a lipase-specific chaperone. Chembiochem, 8(1), 55–60. https://doi.org/10.1002/cbic.200600203.
Binder, U., Matschiner, G., Theobald, I., & Skerra, A. (2010). High-throughput sorting of an anticalin library via EspP-mediated functional display on the Escherichia coli cell surface. Journal of Molecular Biology, 400(4), 783–802. https://doi.org/10.1016/j.jmb.2010.05.049.
Valls, M., de Lorenzo, V., Gonzalez-Duarte, R., & Atrian, S. (2000). Engineering outer-membrane proteins in Pseudomonas putida for enhanced heavy-metal bioadsorption. Journal of Inorganic Biochemistry, 79(1-4), 219–223. https://doi.org/10.1016/S0162-0134(99)00170-1.
Sousa, C., Kotrba, P., Ruml, T., Cebolla, A., & De Lorenzo, V. (1998). Metalloadsorption by Escherichia coli cells displaying yeast and mammalian metallothioneins anchored to the outer membrane protein LamB. Journal of Bacteriology, 180(9), 2280–2284.
Xu, Z., & Lee, S. Y. (1999). Display of polyhistidine peptides on the Escherichia coli cell surface by using outer membrane protein C as an anchoring motif. Applied and Environmental Microbiology, 65(11), 5142–5147.
Taylor, I. M., Harrison, J. L., Timmis, K. N., & O'Connor, C. D. (1990). The TraT lipoprotein as a vehicle for the transport of foreign antigenic determinants to the cell surface of Escherichia Coli K12: structure–function relationships in the TraT protein. Molecular Microbiology, 4(8), 1259–1268. https://doi.org/10.1111/j.1365-2958.1990.tb00705.x.
Francisco, J. A., Campbell, R., Iverson, B. L., & Georgiou, G. (1993). Production and fluorescence-activated cell sorting of Escherichia coli expressing a functional antibody fragment on the external surface. Proceedings of the National Academy of Sciences of the United States of America, 90(22), 10444–10448. https://doi.org/10.1073/pnas.90.22.10444.
Yang, C., Zhao, Q., Liu, Z., Li, Q. Y., Qiao, C. L., Mulchandani, A., & Chen, W. (2008). Cell surface display of functional macromolecule fusions on Escherichia coli for development of an autofluorescent whole-cell biocatalyst. Environmental Science & Technology, 42(16), 6105–6110. https://doi.org/10.1021/es800441t.
Shimazu, M., Mulchandani, A., & Chen, W. (2001). Cell surface display of organophosphorus hydrolase using ice nucleation protein. Biotechnology Progress, 17(1), 76–80. https://doi.org/10.1021/bp0001563.
Kawahara, H. (2002). The structures and functions of ice crystal-controlling proteins from bacteria. Journal of Bioscience and Bioengineering, 94(6), 492–496. https://doi.org/10.1016/S1389-1723(02)80185-2.
Maki, L. R., Galyan, E. L., Changchi, M. M., & Caldwell, D. R. (1974). Ice nucleation induced by Pseudomonas syringae. Applied Microbiology, 28(3), 456–459.
Wu, Z. Q., Qin, L., & Walker, V. K. (2009). Characterization and recombinant expression of a divergent ice nucleation protein from ‘Pseudomonas borealis’. Microbiology-Sgm, 155(4), 1164–1169. https://doi.org/10.1099/mic.0.025114-0.
Rogers, J. S., Stall, R. E., & Burke, M. J. (1987). Low-temperature conditioning of the ice nucleation active bacterium, Erwinia Herbicola. Cryobiology, 24(3), 270–279. https://doi.org/10.1016/0011-2240(87)90030-7.
Kim, H. K., Orser, C., Lindow, S. E., & Sands, D. C. (1987). Xanthomonas-Campestris pv translucens strains active in ice nucleation. Plant Disease, 71(11), 994–997. https://doi.org/10.1094/PD-71-0994.
Koebnik, R., Locher, K. P., & Van Gelder, P. (2000). Structure and function of bacterial outer membrane proteins: barrels in a nutshell. Molecular Microbiology, 37(2), 239–253. https://doi.org/10.1046/j.1365-2958.2000.01983.x.
Wu, M. L., Tsai, C. Y., & Chen, T. H. (2006). Cell surface display of Chi92 on Escherichia coli using ice nucleation protein for improved catalytic and antifungal activity. FEMS Microbiology Letters, 256(1), 119–125. https://doi.org/10.1111/j.1574-6968.2006.00115.x.
Wang, A. J. A., Mulchandani, A., & Chen, W. (2002). Specific adhesion to cellulose and hydrolysis of organophosphate nerve agents by a genetically engineered Escherichia coli strain with a surface-expressed cellulose-binding domain and organophosphorus hydrolase. Applied and Environmental Microbiology, 68(4), 1684–1689. https://doi.org/10.1128/AEM.68.4.1684-1689.2002.
Yang, C., Freudl, R., Qiao, C., & Mulchandani, A. (2010). Cotranslocation of methyl parathion hydrolase to the periplasm and of organophosphorus hydrolase to the cell surface of Escherichia coli by the Tat pathway and ice nucleation protein display system. Applied and Environmental Microbiology, 76(2), 434–440. https://doi.org/10.1128/AEM.02162-09.
Zhang, H. X., Li, Q. Q., Ye, T., Zhang, Z., & Li, L. (2013). Optimization of the whole-cell catalytic activity of recombinant Escherichia coli cells with surface-immobilized organophosphorus hydrolase. Journal of Environmental Biology, 34, 315–319 2 Spec No.
Liang, B., Li, L., Mascin, M., & Liu, A. H. (2012). Construction of xylose dehydrogenase displayed on the surface of bacteria using ice nucleation protein for sensitive d-Xylose detection. Analytical Chemistry, 84(1), 275–282. https://doi.org/10.1021/ac202513u.
Liang, B., Li, L., Tang, X., Lang, Q., Wang, H., Li, F., Shi, J., Shen, W., Palchetti, I., Mascini, M., & Liu, A. (2013). Microbial surface display of glucose dehydrogenase for amperometric glucose biosensor. Biosensors & Bioelectronics, 45, 19–24. https://doi.org/10.1016/j.bios.2013.01.050.
Charbonneau, M. E., & Mourez, M. (2008). The Escherichia coli AIDA-I autotransporter undergoes cytoplasmic glycosylation independently of export. Research in Microbiology, 159(7-8), 537–544. https://doi.org/10.1016/j.resmic.2008.06.009.
Brockmeyer, J., Spelten, S., Kuczius, T., Bielaszewska, M., & Karch, H. (2009). Structure and function relationship of the autotransport and proteolytic activity of EspP from Shiga toxin-producing Escherichia coli. PLoS One, 4(7), e6100. https://doi.org/10.1371/journal.pone.0006100.
Inatsuka, C. S., Xu, Q. A., Vujkovic-Cvijin, I., Wong, S., Stibitz, S., Miller, J. F., & Cotter, P. A. (2010). Pertactin is required for Bordetella species to resist neutrophil-mediated clearance. Infection and Immunity, 78(7), 2901–2909. https://doi.org/10.1128/IAI.00188-10.
Roussel-Jazede, V., Arenas, J., Langereis, J. D., Tommassen, J., & van Ulsen, P. (2014). Variable processing of the IgA protease autotransporter at the cell surface of Neisseria meningitidis. Microbiology-Sgm, 160(Pt_11), 2421–2431. https://doi.org/10.1099/mic.0.082511-0.
Fink, D. L., & St Geme, J. W. (2003). Chromosomal expression of the Haemophilus influenzae Hap autotransporter allows fine-tuned regulation of adhesive potential via inhibition of intermolecular autoproteolysis. Journal of Bacteriology, 185(5), 1608–1615. https://doi.org/10.1128/JB.185.5.1608-1615.2003.
Berg, B. V. D. (2010). Crystal structure of a full-length autotransporter. Journal of Molecular Biology, 396(3), 627–633. https://doi.org/10.1016/j.jmb.2009.12.061.
Wilhelm, S., Rosenau, F., Kolmar, H., & Jaeger, K. E. (2011). Autotransporters with GDSL passenger domains: molecular physiology and biotechnological applications. Chembiochem, 12(10), 1476–1485. https://doi.org/10.1002/cbic.201100013.
Brandon, L. D., & Goldberg, M. B. (2001). Periplasmic transit and disulfide bond formation of the autotransported Shigella protein IcsA. Journal of Bacteriology, 183(3), 951–958. https://doi.org/10.1128/JB.183.3.951-958.2001.
Mistry, D., & Stockley, R. A. (2006). IgA1 protease. International Journal of Biochemistry & Cell Biology, 38(8), 1244–1248. https://doi.org/10.1016/j.biocel.2005.10.005.
Nicolay, T., Vanderleyden, J., & Spaepen, S. (2015). Autotransporter-based cell surface display in Gram-negative bacteria. Critical Reviews in Microbiology, 41(1), 109–123. https://doi.org/10.3109/1040841X.2013.804032.
Jose, J., & Meyer, T. F. (2007). The autodisplay story, from discovery to biotechnical and biomedical applications. Microbiology and Molecular Biology Reviews, 71(4), 600–619. https://doi.org/10.1128/MMBR.00011-07.
Schneewind, O., Model, P., & Fischetti, V. A. (1992). Sorting of protein A to the staphylococcal cell-wall. Cell, 70(2), 267–281. https://doi.org/10.1016/0092-8674(92)90101-H.
Schneewind, O., Fowler, A., & Faull, K. F. (1995). Structure of the cell-wall anchor of surface-proteins in Staphylococcus aureus. Science, 268(5207), 103–106. https://doi.org/10.1126/science.7701329.
Stahl, S., Robert, A., Gunneriusson, E., Wernerus, H., Cano, F., Liljeqvist, S., Hansson, M., Nguyen, T. N., & Samuelson, P. (2000). Staphylococcal surface display and its applications. International Journal of Medical Microbiology, 290(7), 571–577. https://doi.org/10.1016/S1438-4221(00)80002-8.
Sleytr, U. B., & Sara, M. (1997). Bacterial and archaeal S-layer proteins: Structure-function relationships and their biotechnological applications. Trends in Biotechnology, 15(1), 20–26. https://doi.org/10.1016/S0167-7799(96)10063-9.
Boot, H. J., & Pouwels, P. H. (1996). Expression, secretion and antigenic variation of bacterial S-layer proteins. Molecular Microbiology, 21(6), 1117–1123. https://doi.org/10.1046/j.1365-2958.1996.711442.x.
Ribeiro, L. A., Azevedo, V., Le Loir, Y., Oliveira, S. C., Dieye, Y., Piard, J. C., Gruss, A., & Langella, P. (2002). Production and targeting of the Brucella abortus antigen L7/L12 in Lactococcus lactis: a first step towards food-grade live vaccines against brucellosis. Applied and Environmental Microbiology, 68(2), 910–916. https://doi.org/10.1128/AEM.68.2.910-916.2002.
Potot, S., Serra, C. R., Henriques, A. O., & Schyns, G. (2010). Display of recombinant proteins on Bacillus subtilis spores, using a coat-associated enzyme as the carrier. Applied and Environmental Microbiology, 76(17), 5926–5933. https://doi.org/10.1128/AEM.01103-10.
Hinc, K., Isticato, R., Dembek, M., Karczewska, J., Iwanicki, A., Peszynska-Sularz, G., De Felice, M., Obuchowski, M., & Ricca, E. (2010). Expression and display of UreA of Helicobacter acinonychis on the surface of Bacillus subtilis spores. Microbial Cell Factories, 9(1), 2. https://doi.org/10.1186/1475-2859-9-2.
Wang, N., Chang, C., Yao, Q., Li, G. H., Qin, L. G., Chen, L., & Chen, K. P. (2011). Display of Bombyx mori alcohol dehydrogenases on the Bacillus subtilis spore surface to enhance enzymatic activity under adverse conditions. PLoS One, 6(6), e21454. https://doi.org/10.1371/journal.pone.0021454.
Negri, A., Potocki, W., Iwanicki, A., Obuchowski, M., & Hinc, K. (2013). Expression and display of Clostridium difficile protein FliD on the surface of Bacillus subtilis spores. Journal of Medical Microbiology, 62(Pt_9), 1379–1385. https://doi.org/10.1099/jmm.0.057372-0.
Valdez, A., Yepiz-Plascencia, G., Ricca, E., & Olmos, J. (2014). First Litopenaeus vannamei WSSV 100% oral vaccination protection using CotC::Vp26 fusion protein displayed on Bacillus subtilis spores surface. Journal of Applied Microbiology, 117(2), 347–357. https://doi.org/10.1111/jam.12550.
Lian, C. Q., Zhou, Y., Feng, F., Chen, L., Tang, Q., Yao, Q., & Chen, K. P. (2014). Surface display of human growth hormone on Bacillus subtilis spores for oral administration. Current Microbiology, 68(4), 463–471. https://doi.org/10.1007/s00284-013-0500-9.
Nguyen, Q. A., & Schumann, W. (2014). Use of IPTG-inducible promoters for anchoring recombinant proteins on the Bacillus subtilis spore surface. Protein Expression and Purification, 95, 67–76. https://doi.org/10.1016/j.pep.2013.11.018.
Chen, H. Y., Tian, R., Ni, Z., Zhang, Q., Zhang, T. X., Chen, Z., Chen, K. P., & Yang, S. L. (2015). Surface display of the thermophilic lipase Tm1350 on the spore of Bacillus subtilis by the CotB anchor protein. Extremophiles, 19(4), 799–808. https://doi.org/10.1007/s00792-015-0755-0.
Chen, H. Y., Zhang, T. X., Jia, J. R., Vastermark, A., Tian, R., Ni, Z., Chen, Z., Chen, K. P., & Yang, S. L. (2015). Expression and display of a novel thermostable esterase from Clostridium thermocellum on the surface of Bacillus subtilis using the CotB anchor protein. Journal of Industrial Microbiology & Biotechnology, 42(11), 1439–1448. https://doi.org/10.1007/s10295-015-1676-8.
Isticato, R., Cangiano, G., Tran, H. T., Ciabattini, A., Medaglini, D., Oggioni, M. R., De Felice, M., Pozzi, G., & Ricca, E. (2001). Surface display of recombinant proteins on Bacillus subtilis spores. Journal of Bacteriology, 183(21), 6294–6301. https://doi.org/10.1128/JB.183.21.6294-6301.2001.
Isticato, R., Di Mase, D. S., Mauriello, E. M. F., De Felice, M., & Ricca, E. (2007). Amino terminal fusion of heterologous proteins to CotC increases display efficiencies in the Bacillus subtilis spore system. BioTechniques, 42(2), 151–156. https://doi.org/10.2144/000112329.
Fujita, Y., Takahashi, S., Ueda, M., Tanaka, A., Okada, H., Morikawa, Y., Kawaguchi, T., Arai, M., Fukuda, H., & Kondo, A. (2002). Direct and efficient production of ethanol from cellulosic material with a yeast strain displaying cellulolytic enzymes. Applied and Environmental Microbiology, 68(10), 5136–5141. https://doi.org/10.1128/AEM.68.10.5136-5141.2002.
Zou, W., Ueda, M., & Tanaka, A. (2002). Screening of a molecule endowing Saccharomyces cerevisiae with n-nonane-tolerance from a combinatorial random protein library. Applied Microbiology and Biotechnology, 58(6), 806–812. https://doi.org/10.1007/s00253-002-0961-4.
Kuroda, K., & Ueda, M. (2006). Effective display of metallothionein tandem repeats on the bioadsorption of cadmium ion. Applied Microbiology and Biotechnology, 70(4), 458–463. https://doi.org/10.1007/s00253-005-0093-8.
Wang, Z. K., Qi, Q. S., & Wang, P. G. (2006). Engineering of cyclodextrin glucanotransferase on the cell surface of Saccharomyces cerevisiae for improved cyclodextrin production. Applied and Environmental Microbiology, 72(3), 1873–1877. https://doi.org/10.1128/AEM.72.3.1873-1877.2006.
Jin, M., Song, G., Carman, C. V., Kim, Y. S., Astrof, N. S., Shimaoka, M., Wittrup, K. D., & Springer, T. A. (2006). Directed evolution to probe protein allostery and integrin I domains of 200,000-fold higher affinity. Proceedings of the National Academy of Sciences of the United States of America, 103(15), 5758–5763. https://doi.org/10.1073/pnas.0601164103.
Sato, N., Matsumoto, T., Ueda, M., Tanaka, A., Fukuda, H., & Kondo, A. (2002). Long anchor using Flo1 protein enhances reactivity of cell surface-displayed glucoamylase to polymer substrates. Applied Microbiology and Biotechnology, 60(4), 469–474. https://doi.org/10.1007/s00253-002-1121-6.
Matsumoto, T., Fukuda, H., Ueda, M., Tanaka, A., & Kondo, A. (2002). Construction of yeast strains with high cell surface lipase activity by using novel display systems based on the Flo1p flocculation functional domain. Applied and Environmental Microbiology, 68(9), 4517–4522. https://doi.org/10.1128/AEM.68.9.4517-4522.2002.
Abe, H., Ohba, M., Shimma, Y., & Jigami, Y. (2004). Yeast cells harboring human alpha-1,3-fucosyltransferase at the cell surface engineered using Pir, a cell wall-anchored protein. FEMS Yeast Research, 4(4-5), 417–425. https://doi.org/10.1016/S1567-1356(03)00193-4.
Abe, H., Shimma, Y., & Jigami, Y. (2003). In vitro oligosaccharide synthesis using intact yeast cells that display glycosyltransferases at the cell surface through cell wall-anchored protein Pir. Glycobiology, 13(2), 87–95. https://doi.org/10.1093/glycob/cwg014.
Andres, I., Gallardo, O., Parascandola, P., Pastor, F. I. J., & Zueco, J. (2005). Use of the cell wall protein pir4 as a fusion partner for the expression of Bacillus sp BP-7 xylanase A in Saccharomyces cerevisiae. Biotechnology and Bioengineering, 89(6), 690–697. https://doi.org/10.1002/bit.20375.
Kato, M., Kuzuhara, Y., Maeda, H., Shiraga, S., & Ueda, M. (2006). Analysis of a processing system for proteases using yeast cell surface engineering: conversion of precursor of proteinase A to active proteinase A. Applied Microbiology and Biotechnology, 72(6), 1229–1237. https://doi.org/10.1007/s00253-006-0408-4.
Breinig, F., Diehl, B., Rau, S., Zimmer, C., Schwab, H., & Schmitt, M. J. (2006). Cell surface expression of bacterial esterase A by Saccharomyces cerevisiae and its enhancement by constitutive activation of the cellular unfolded protein response. Applied and Environmental Microbiology, 72(11), 7140–7147. https://doi.org/10.1128/AEM.00503-06.
Zhao, H., Shen, Z. M., Kahn, P. C., & Lipke, P. N. (2001). Interaction of alpha-agglutinin and a-agglutinin, Saccharomyces cerevisiae sexual cell adhesion molecules. Journal of Bacteriology, 183(9), 2874–2880. https://doi.org/10.1128/JB.183.9.2874-2880.2001.
Verstrepen, K. J., Derdelinckx, G., Verachtert, H., & Delvaux, F. R. (2003). Yeast flocculation: what brewers should know. Applied Microbiology and Biotechnology, 61(3), 197–205. https://doi.org/10.1007/s00253-002-1200-8.
Ecker, M., Deutzmann, R., Lehle, L., Mrsa, V., & Tanner, W. (2006). Pir proteins of Saccharomyces cerevisiae are attached to beta-1,3-glucan by a new protein-carbohydrate linkage. Journal of Biological Chemistry, 281(17), 11523–11529. https://doi.org/10.1074/jbc.M600314200.
Richins, R. D., Kaneva, I., Mulchandani, A., & Chen, W. (1997). Biodegradation of organophosphorus pesticides by surface-expressed organophosphorus hydrolase. Nature Biotechnology, 15(10), 984–987. https://doi.org/10.1038/nbt1097-984.
Shimazu, M., Nguyen, A., Mulchandani, A., & Chen, W. (2003). Cell surface display of organophosphorus hydrolase in Pseudomonas putida using an ice-nucleation protein anchor. Biotechnology Progress, 19(5), 1612–1614. https://doi.org/10.1021/bp0340640.
Takayama, K., Suye, S., Kuroda, K., Ueda, M., Kitaguchi, T., Tsuchiyama, K., Fukuda, T., Chen, W., & Mulchandani, A. (2006). Surface display of organophosphorus hydrolase on Saccharomyces cerevisiae. Biotechnology Progress, 22(4), 939–943. https://doi.org/10.1021/bp060107b.
Han, L., & Liu, A. (2017). Novel cell–inorganic hybrid catalytic interfaces with enhanced enzymatic activity and stability for sensitive biosensing of paraoxon. ACS Applied Materials & Interfaces, 9(8), 6894–6901. https://doi.org/10.1021/acsami.6b15992.
Lee, S. H., Lee, S. Y., & Park, B. C. (2005). Cell surface display of lipase in Pseudomonas putida KT2442 using OprF as an anchoring motif and its biocatalytic applications. Applied and Environmental Microbiology, 71(12), 8581–8586. https://doi.org/10.1128/AEM.71.12.8581-8586.2005.
Lee, S. H., Choi, J. I., Park, S. J., Lee, S. Y., & Park, B. C. (2004). Display of bacterial lipase on the Escherichia coli cell surface by using FadL as an anchoring motif and use of the enzyme in enantioselective biocatalysis. Applied and Environmental Microbiology, 70(9), 5074–5080. https://doi.org/10.1128/AEM.70.9.5074-5080.2004.
Becker, S., Theile, S., Heppeler, N., Michalczyk, A., Wentzel, A., Wilhelm, S., Jaeger, K. E., & Kolmar, H. (2005). A generic system for the Escherichia coli cell-surface display of lipolytic enzymes. FEBS Letters, 579(5), 1177–1182. https://doi.org/10.1016/j.febslet.2004.12.087.
Liu, Y., Zhang, R., Lian, Z., Wang, S., & Wright, A. T. (2014). Yeast cell surface display for lipase whole cell catalyst and its applications. Journal of Molecular Catalysis B: Enzymatic, 106, 17–25. https://doi.org/10.1016/j.molcatb.2014.04.011.
Moura, M. V. H., da Silva, G. P., Machado, A. C. D., Torres, F. A. G., Freire, D. M. G., & Almeida, R. V. (2015). Displaying lipase B from Candida antarctica in Pichia pastoris using the yeast surface display approach: prospection of a new anchor and characterization of the whole cell biocatalyst. PLoS One, 10(10), e0141454. https://doi.org/10.1371/journal.pone.0141454.
Yuzbasheva, E. Y., Yuzbashev, T. V., Perkovskaya, N. I., Mostova, E. B., Vybornaya, T. V., Sukhozhenko, A. V., Toropygin, I. Y., & Sineoky, S. P. (2015). Cell surface display of Yarrowia lipolytica lipase lip2p using the cell wall protein YlPir1p, its characterization, and application as a whole-cell biocatalyst. Applied Biochemistry and Biotechnology, 175(8), 3888–3900. https://doi.org/10.1007/s12010-015-1557-7.
Chen, X. Z. (2017). Yeast cell surface display: an efficient strategy for improvement of bioethanol fermentation performance. Bioengineered, 8(2), 115–119. https://doi.org/10.1080/21655979.2016.1212135.
Ito, J., Fujita, Y., Ueda, M., Fukuda, H., & Kondo, A. (2004). Improvement of cellulose-degrading ability of a yeast strain displaying Trichoderma reesei endoglucanase II by recombination of cellulose-binding domains. Biotechnology Progress, 20(3), 688–691. https://doi.org/10.1021/bp034332u.
Kaya, M., Ito, J., Kotaka, A., Matsumura, K., Bando, H., Sahara, H., Ogino, C., Shibasaki, S., Kuroda, K., Ueda, M., Kondo, A., & Hata, Y. (2008). Isoflavone aglycones production from isoflavone glycosides by display of beta-glucosidase from Aspergillus oryzae on yeast cell surface. Applied Microbiology and Biotechnology, 79(1), 51–60. https://doi.org/10.1007/s00253-008-1393-6.
Boder, E. T., & Wittrup, K. D. (1997). Yeast surface display for screening combinatorial polypeptide libraries. Nature Biotechnology, 15(6), 553–557. https://doi.org/10.1038/nbt0697-553.
Katahira, S., Fujita, Y., Mizuike, A., Fukuda, H., & Kondo, A. (2004). Construction of a xylan-fermenting yeast strain through codisplay of xylanolytic enzymes on the surface of xylose-utilizing Saccharomyces cerevisiae cells. Applied and Environmental Microbiology, 70(9), 5407–5414. https://doi.org/10.1128/AEM.70.9.5407-5414.2004.
Khaw, T. S., Katakura, Y., Koh, J., Kondo, A., Ueda, M., & Shioya, S. (2006). Evaluation of performance of different surface-engineered yeast strains for direct ethanol production from raw starch. Applied Microbiology and Biotechnology, 70(5), 573–579. https://doi.org/10.1007/s00253-005-0101-z.
Tsai, S. L., Oh, J., Singh, S., Chen, R. Z., & Chen, W. (2009). Functional assembly of minicellulosomes on the Saccharomyces cerevisiae cell surface for cellulose hydrolysis and ethanol production. Applied and Environmental Microbiology, 75(19), 6087–6093. https://doi.org/10.1128/AEM.01538-09.
Goyal, G., Tsai, S. L., Madan, B., DaSilva, N. A., & Chen, W. (2011). Simultaneous cell growth and ethanol production from cellulose by an engineered yeast consortium displaying a functional mini-cellulosome. Microbial Cell Factories, 10(1), 89–96. https://doi.org/10.1186/1475-2859-10-89.
Tsai, S. L., DaSilva, N. A., & Chen, W. (2013). Functional display of complex cellulosomes on the yeast surface via adaptive assembly. ACS Synthetic Biology, 2(1), 14–21. https://doi.org/10.1021/sb300047u.
Wen, F., Sun, J., & Zhao, H. M. (2010). Yeast surface display of trifunctional minicellulosomes for simultaneous saccharification and fermentation of cellulose to ethanol. Applied and Environmental Microbiology, 76(4), 1251–1260. https://doi.org/10.1128/AEM.01687-09.
Fan, L. H., Zhang, Z. J., Yu, X. Y., Xue, Y. X., & Tan, T. W. (2012). Self-surface assembly of cellulosomes with two miniscaffoldins on Saccharomyces cerevisiae for cellulosic ethanol production. Proceedings of the National Academy of Sciences of the United States of America, 109(33), 13260–13265. https://doi.org/10.1073/pnas.1209856109.
Feng, R. R., Liang, B., Hou, C. T., Han, D. F., Han, L., Lang, Q. L., Liu, A. H., & Han, L. H. (2016). Rational design of xylose dehydrogenase for improved thermostability and its application in development of efficient enzymatic biofuel cell. Enzyme and Microbial Technology, 84, 78–85. https://doi.org/10.1016/j.enzmictec.2015.12.002.
Tang, X., Zhang, T., Liang, B., Han, D., Zeng, L., Zheng, C., Li, T., Wei, M., & Liu, A. (2014). Sensitive electrochemical microbial biosensor for p-nitrophenylorganophosphates based on electrode modified with cell surface-displayed organophosphorus hydrolase and ordered mesopore carbons. Biosensors and Bioelectronics, 60, 137–142. https://doi.org/10.1016/j.bios.2014.04.001.
Liu, R. H., Yang, C., Xu, Y. M., Xu, P., Jiang, H., & Qiao, C. L. (2013). Development of a whole-cell biocatalyst/biosensor by display of multiple heterologous proteins on the Escherichia coli cell surface for the detoxification and detection of organophosphates. Journal of Agricultural and Food Chemistry, 61(32), 7810–7816. https://doi.org/10.1021/jf402999b.
Li, J. Q., Ba, Q., Yin, J., Wu, S. J., Zhuan, F. F., Xu, S. C., Li, J. Y., Salazar, J. K., Zhang, W., & Wang, H. (2013). Surface display of recombinant Drosophila melanogaster acetylcholinesterase for detection of organic phosphorus and carbamate pesticides. PLoS One, 8(9), e72986. https://doi.org/10.1371/journal.pone.0072986.
Matsuura, H., Yamamoto, Y., Muraoka, M., Akaishi, K., Hori, Y., Uemura, K., Tsuji, N., Harada, K., Hirata, K., Bamba, T., Miyasaka, H., Kuroda, K., & Ueda, M. (2013). Development of surface-engineered yeast cells displaying phytochelatin synthase and their application to cadmium biosensors by the combined use of pyrene-excimer fluorescence. Biotechnology Progress, 29(5), 1197–1202. https://doi.org/10.1002/btpr.1789.
Furst, A. L., Hoepker, A. C., & Francis, M. B. (2017). Quantifying hormone disruptors with an engineered bacterial biosensor. Acs Central Science, 3(2), 110–116. https://doi.org/10.1021/acscentsci.6b00322.
Venkatesh, A. G., Sun, A., Brickner, H., Looney, D., Hall, D. A., & Aronoff-Spencer, E. (2015). Yeast dual-affinity biobricks: progress towards renewable whole-cell biosensors. Biosensors & Bioelectronics, 70, 462–468. https://doi.org/10.1016/j.bios.2015.03.044.
Grewal, Y. S., Shiddiky, M. J. A., Mahler, S. M., Cangelosi, G. A., & Trau, M. (2016). Nanoyeast and other cell envelope compositions for protein studies and biosensor applications. ACS Applied Materials & Interfaces, 8(45), 30649–30664. https://doi.org/10.1021/acsami.6b09263.
Tafakori, V., Torktaz, I., Doostmohammadi, M., & Ahmadian, G. (2012). Microbial cell surface display; its medical and environmental applications. Iranian Journal of Biotechnology, 10, 231–239.
Han, L., Liu, P., Zhang, H., Li, F., & Liu, A. (2017). Phage capsid protein-directed MnO2 nanosheets with peroxidase-like activity for spectrometric biosensing and evaluation of antioxidant behaviour. Chemical Communications, 53(37), 5216–5219. https://doi.org/10.1039/C7CC02049J.
Han, L., Shao, C., Liang, B., & Liu, A. (2016). Genetically engineered phage-templated MnO2 nanowires: synthesis and their application in electrochemical glucose biosensor operated at neutral pH condition. ACS Applied Materials & Interfaces, 8(22), 13768–13776. https://doi.org/10.1021/acsami.6b03266.
Han, L., Yang, D.-P., & Liu, A. (2015). Leaf-templated synthesis of 3D hierarchical porous cobalt oxide nanostructure as direct electrochemical biosensing interface with enhanced electrocatalysis. Biosensors and Bioelectronics, 63, 145–152. https://doi.org/10.1016/j.bios.2014.07.031.
Han, L., Zhang, S., Han, L., Yang, D.-P., Hou, C., & Liu, A. (2014). Porous gold cluster film prepared from Au@BSA microspheres for electrochemical nonenzymatic glucose sensor. Electrochimica Acta, 138, 109–114. https://doi.org/10.1016/j.electacta.2014.06.095.
Han, L., Li, C., Zhang, T., Lang, Q., & Liu, A. (2015). Au@Ag heterogeneous nanorods as nanozyme interfaces with peroxidase-like activity and their application for one-pot analysis of glucose at nearly neutral pH. ACS Applied Materials & Interfaces, 7(26), 14463–14470. https://doi.org/10.1021/acsami.5b03591.
Han, L., Zeng, L., Wei, M., Li, C. M., & Liu, A. (2015). A V2O3-ordered mesoporous carbon composite with novel peroxidase-like activity towards the glucose colorimetric assay. Nanoscale, 7(27), 11678–11685. https://doi.org/10.1039/C5NR02694F.
Funding
This work was supported by National Natural Science Foundation of China (Grant No. 21705087) and Distinguished Scholars of Qingdao Agricultural University (No. 6631117015).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Han, L., Zhao, Y., Cui, S. et al. Redesigning of Microbial Cell Surface and Its Application to Whole-Cell Biocatalysis and Biosensors. Appl Biochem Biotechnol 185, 396–418 (2018). https://doi.org/10.1007/s12010-017-2662-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-017-2662-6