Abstract
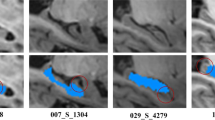
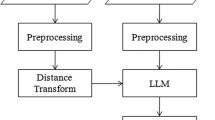
Automatic and reliable segmentation of hippocampus from MR brain images is of great importance in studies of neurological diseases, such as epilepsy and Alzheimer’s disease. In this paper, we proposed a novel metric learning method to fuse segmentation labels in multi-atlas based image segmentation. Different from current label fusion methods that typically adopt a predefined distance metric model to compute a similarity measure between image patches of atlas images and the image to be segmented, we learn a distance metric model from the atlases to keep image patches of the same structure close to each other while those of different structures are separated. The learned distance metric model is then used to compute the similarity measure between image patches in the label fusion. The proposed method has been validated for segmenting hippocampus based on the EADC-ADNI dataset with manually labelled hippocampus of 100 subjects. The experiment results demonstrated that our method achieved statistically significant improvement in segmentation accuracy, compared with state-of-the-art multi-atlas image segmentation methods.




Similar content being viewed by others
References
Akhondi-Asl, A., Jafari-Khouzani, K., Elisevich, K., & Soltanian-Zadeh, H. (2011). Hippocampal volumetry for lateralization of temporal lobe epilepsy: automated versus manual methods. NeuroImage, 54, S218–S226.
Aljabar, P., Heckemann, R., Hammers, A., Hajnal, J., & Rueckert, D. (2009). Multi-atlas based segmentation of brain images: Atlas selection and its effect on accuracy. NeuroImage, 46, 726–738.
Artaechevarria, X., Munoz-Barrutia, A., & Ortiz-de-Solorzano, C. (2009). Combination strategies in multi-atlas image segmentation: Application to brain MR data. IEEE Transactions on Medical Imaging, 28, 1266–1277.
Avants, B. B., Epstein, C. L., Grossman, M., & Gee, J. C. (2008). Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Medical Image Analysis, 12, 26–41.
Bai, W., Shi, W., Ledig, C., & Rueckert, D. (2015). Multi-atlas segmentation with augmented features for cardiac MR images. Medical Image Analysis, 19, 98–109.
Boccardi, M., Bocchetta, M., Morency, F. C., Collins, D. L., Nishikawa, M., Ganzola, R., Grothe, M. J., Wolf, D., Redolfi, A., & Pievani, M. (2015). Training labels for hippocampal segmentation based on the EADC-ADNI harmonized hippocampal protocol. Alzheimer's & Dementia, 11, 175–183.
Carmichael, O. T., Aizenstein, H. A., Davis, S. W., Becker, J. T., Thompson, P. M., Meltzer, C. C., & Liu, Y. (2005). Atlas-based hippocampus segmentation in Alzheimer’s disease and mild cognitive impairment. NeuroImage, 27, 979–990.
Chang, C. C., & Lin, C. J. (2011). LIBSVM: A library for support vector machines. ACM Transactions on Intelligent Systems and Technology (TIST), 2(3), 27.
Cheng, H., & Fan, Y. (2014). Functional parcellation of the hippocampus by clustering resting state fMRI signals. In: 2014 I.E. 11th International Symposium on Biomedical Imaging (ISBI), pp 5–8.
Chupin, M., Mukuna-Bantumbakulu, A. R., Hasboun, D., Bardinet, E., Baillet, S., Kinkingnéhun, S., Lemieux, L., Dubois, B., & Garnero, L. (2007). Anatomically constrained region deformation for the automated segmentation of the hippocampus and the amygdala: method and validation on controls and patients with Alzheimer’s disease. NeuroImage, 34, 996–1019.
Coupé, P., Manjón, J. V., Fonov, V., Pruessner, J., Robles, M., & Collins, D. L. (2011). Patch-based segmentation using expert priors: application to hippocampus and ventricle segmentation. NeuroImage, 54, 940–954.
den Heijer, T., van der Lijn, F., Vernooij, M. W., de Groot, M., Koudstaal, P., van der Lugt, A., Krestin, G. P., Hofman, A., Niessen, W. J., & Breteler, M. M. (2012). Structural and diffusion MRI measures of the hippocampus and memory performance. NeuroImage, 63, 1782–1789.
Dill, V., Franco, A. R., & Pinho, M. S. (2015). Automated methods for hippocampus segmentation: the evolution and a review of the state of the art. Neuroinformatics, 13, 133–150.
Doshi, J., Erus, G., Ou, Y., Resnick, S. M., Gur, R. C., Gur, R. E., Satterthwaite, T. D., Furth, S., & Davatzikos, C. (2016). MUSE: MUlti-atlas region Segmentation utilizing Ensembles of registration algorithms and parameters, and locally optimal atlas selection. NeuroImage, 127, 186–195.
Giraud, R., Ta, V.-T., Papadakis, N., Manjón, J. V., Collins, D. L., Coupé, P., & Initiative, A. D. N. (2016). An Optimized PatchMatch for multi-scale and multi-feature label fusion. NeuroImage, 124, 770–782.
Guillaumin M, Verbeek J, Schmid C (2009) Is that you? Metric learning approaches for face identification. In: Computer Vision, 2009 I.E. 12th International Conference on, pp 498–505: IEEE.
Hao, Y., Jiang, T., & Fan, Y. (2012a). Shape-constrained multi-atlas based segmentation with multichannel registration. SPIE Medical Imaging. International Society for Optics and Photonics, pp. 83143N-83143N-83148.
Hao, Y., Liu, J., Duan, Y., Zhang, X., Yu, C., Jiang, T., & Fan, Y. (2012b). Local label learning (L3) for multi-atlas based segmentation. SPIE Medical Imaging. International Society for Optics and Photonics, pp. 83142E-83142E-83148.
Hao, Y., Wang, T., Zhang, X., Duan, Y., Yu, C., Jiang, T., & Fan, Y. (2014). Local label learning (LLL) for subcortical structure segmentation: Application to hippocampus segmentation. Human Brain Mapping, 35, 2674–2697.
Heckemann, R. A., Hajnal, J. V., Aljabar, P., Rueckert, D., & Hammers, A. (2006). Automatic anatomical brain MRI segmentation combining label propagation and decision fusion. NeuroImage, 33, 115–126.
Iglesias, J. E., & Sabuncu, M. R. (2015). Multi-atlas segmentation of biomedical images: A survey. Medical Image Analysis, 24, 205–219.
Jafari-Khouzani, K., Elisevich, K. V., Patel, S., & Soltanian-Zadeh, H. (2011). Dataset of magnetic resonance images of nonepileptic subjects and temporal lobe epilepsy patients for validation of hippocampal segmentation techniques. Neuroinformatics, 9, 335–346.
Liao, S., Gao, Y., Lian, J., & Shen, D. (2013). Sparse patch-based label propagation for accurate prostate localization in CT images. IEEE Transactions on Medical Imaging, 32, 419–434.
Lötjönen, J. M. P., Wolz, R., Koikkalainen, J. R., Thurfjell, L., Waldemar, G., Soininen, H., & Rueckert, D. (2010). Fast and robust multi-atlas segmentation of brain magnetic resonance images. NeuroImage, 49, 2352–2365.
Rohlfing, T., Brandt, R., Menzel, R., & Maurer Jr., C. R. (2004). Evaluation of atlas selection strategies for atlas-based image segmentation with application to confocal microscopy images of bee brains. NeuroImage, 21, 1428–1442.
Rousseau, F., Habas, P. A., & Studholme, C. (2011). A supervised patch-based approach for human brain labeling. IEEE Transactions on Medical Imaging, 30, 1852–1862.
Sabuncu, M. R., Yeo, B. T. T., Van Leemput, K., Fischl, B., & Golland, P. (2010). A generative model for image segmentation based on label fusion. IEEE Transactions on Medical Imaging, 29, 1714–1729.
Sanroma, G., Wu, G., Gao, Y., Thung, K.-H., Guo, Y., & Shen, D. (2015). A transversal approach for patch-based label fusion via matrix completion. Medical Image Analysis, 24, 135–148.
Tong, T., Wolz, R., Wang, Z., Gao, Q., Misawa, K., Fujiwara, M., Mori, K., Hajnal, J. V., & Rueckert, D. (2015). Discriminative dictionary learning for abdominal multi-organ segmentation. Medical Image Analysis, 23, 92–104.
Wang, H., Suh, J. W., Das, S. R., Pluta, J. B., Craige, C., & Yushkevich, P. A. (2013). Multi-atlas segmentation with joint label fusion. IEEE Transactions on Pattern Analysis and Machine Intelligence, 35, 611–623.
Wang, H., Cao, Y., & Syeda-Mahmood, T. (2014). Multi-atlas segmentation with learning-based label fusion. Machine learning in Medical Imaging, 256–263.
Wang, F., Zuo, W., Zhang, L., Meng, D., & Zhang, D. (2015). A kernel classification framework for metric learning. IEEE Transactions on Neural Networks and Learning Systems, 26, 1950–1962.
Warfield, S. K., Zou, K. H., & Wells, W. M. (2004). Simultaneous truth and performance level estimation (STAPLE): an algorithm for the validation of image segmentation. IEEE Transactions on Medical Imaging, 23, 903–921.
Weinberger, K. Q., & Saul, L. K. (2009). Distance metric learning for large margin nearest neighbor classification. Journal of Machine Learning Research, 10, 207–244.
Wolz, R., Schwarz, A. J., Yu, P., Cole, P. E., Rueckert, D., Jack, C. R., Raunig, D., Hill, D., & Initiative AsDN (2014). Robustness of automated hippocampal volumetry across magnetic resonance field strengths and repeat images. Alzheimer's & Dementia, 10, 430–438 e432.
Wu, Y., Liu, G., Huang, M., Guo, J., Jiang, J., Yang, W., Chen, W., & Feng, Q. (2014). Prostate segmentation based on variant scale patch and local independent projection. IEEE Transactions on Medical Imaging, 33, 1290–1303.
Wu, G., Kim, M., Sanroma, G., Wang, Q., Munsell, B. C., Shen, D., & Initiative, A. D. N. (2015). Hierarchical multi-atlas label fusion with multi-scale feature representation and label-specific patch partition. NeuroImage, 106, 34–46.
Xie, Q., & Ruan, D. (2014). Low-complexity atlas-based prostate segmentation by combining global, regional, and local metrics. Medical Physics, 41, 041909.
Xing, E. P., Jordan, M. I., Russell, S., & Ng, A. Y. (2002). Distance metric learning with application to clustering with side-information. In: Advances in Neural Information Processing Systems, pp 505–512.
Yan, P.-g., Cao, Y., Yuan, Y., Turkbey, B., & Choyke, P. L. (2015). Label Image Constrained Multiatlas Selection. IEEE transactions on Cybernetics, 45, 1158–1168.
Zhu, H., Cheng, H., & Fan, Y. (2015). Random local binary pattern based label learning for multi-atlas segmentation. SPIE Medical Imaging. International Society for Optics and Photonics, pp. 94131B-94131B-94138.
Acknowledgments
This work was supported in part by National Key Basic Research and Development Program (No. 2015CB856404), National Natural Science Foundation of China (No. 81271514, 61473296, 61602307), and NIH grants EB022573, CA189523, and AG014971.
Author information
Authors and Affiliations
Consortia
Corresponding author
Additional information
Alzheimer’s Disease Neuroimaging Initiative (ADNI) is a Group/Institutional Author.
Data used in preparation of this article were obtained from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database (adni.loni.ucla.edu). As such, the investigators within the ADNI contributed to the design and implementation of ADNI and/or provided data but did not participate in analysis or writing of this report. A complete listing of ADNI investigators can be found at: http://adni.loni.ucla.edu/wp-content/uploads/how_to_apply/ADNI_Acknowledgement_List.pdf
Rights and permissions
About this article
Cite this article
Zhu, H., Cheng, H., Yang, X. et al. Metric Learning for Multi-atlas based Segmentation of Hippocampus. Neuroinform 15, 41–50 (2017). https://doi.org/10.1007/s12021-016-9312-y
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12021-016-9312-y