Abstract
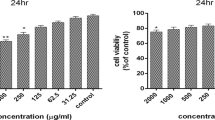
Apoptosis is induced in MCF-7 breast cancer cells following treatment with salicylic acid (20 mM), either in the presence or absence of a heat shock (42°C for 30 min). In order to study the alterations of apoptotic genes with quantitative real-time PCR (qPCR), suitable genes with unchanged expression following the treatments is required for normalizing the gene expression levels. In this study, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), β-actin (ACTB), Histone H2A (HIST), constitutively expressed heat shock protein 70 (HSC70) and tyrosine 3-monooxygenase/trytophan 5 monooxygenase activation protein, 14-3-3 (YWHAZ) were evaluated as appropriate reference genes. Analysis of gene expression data with one-way ANOVA, geNorm and NormFinder identified HIST and YWHAZ as the least affected during the induction of apoptosis by the different treatments, and is the most suitable gene-pair for normalization during qPCR analysis in MCF-7 breast cancer cells undergoing apoptosis following treatment with SA and/or HS.




Similar content being viewed by others
References
Kerr, J. F., Wylie, A. H., & Currie, A. R. (1972). Apoptosis: A basic biological phenomenon with wide-ranging implications in tissue kinetics. British Journal of Cancer, 26(4), 239–257.
Elmore, S. (2007). Apoptosis: A review of programmed cell death. Toxicologic Pathology, 35, 495–516.
Kumar, S. (2007). Caspase function in programmed cell death. Cell Death and Differentiation, 14, 32–43.
Galluzzi, L., Aaronson, S. A., Abrahms, J., Alnemri, E. S., Andrews, D. W., Baehrecke, E. H., et al. (2009). Guidelines for the use and interpretation of assays for monitoring cell death in higher eukaryotes. Cell Death and Differentiation, 16, 1093–1107.
Bustin, S. A. (2002). Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): Trends and problems. Journal of Molecular Endocrinology, 29, 23–39.
Bustin, S. A. (2010). Why the need for qPCR publication guidelines?—The case for MIQE. Methods, 50, 217–226.
Derveaux, S., Vandesompele, J., & Hellemans, J. (2010). How to do successful gene expression analysis using real-time PCR. Methods, 50, 227–230.
Thellin, O., Zorzi, W., Lakaye, B., De Borman, B., Coumans, B., & Hennen, G. (1999). Housekeeping genes as internal standards: Use and limits. Journal of Biotechnology, 75, 291–295.
Bustin, S. A., & Nolan, T. (2004). Pitfalls of quantitative real-time reverse transcription polymerase chain reaction. Journal of Biomolecular Techniques, 15, 155–166.
Taylor, S., Wakem, M., Dijkman, G., Alsarraj, M., & Nguyen, M. (2010). A practical approach to RT-qPCR—publishing data that conform to the MIQE guidelines. Methods, 50, S1–S5.
Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A., et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology, 3(7), 34.1–34.11.
Erickson, H. S., Albert, P. S., Gillespie, J. W., Wallis, B. S., Rodriguez-Canales, J., Linehan, W. M., et al. (2007). Assessment of normalization strategies for quantitative RT-PCR using microdissected tissue samples. Laboratory Investigation, 87, 951–962.
Suzuki, T., Higgins, P. J., & Crawford, D. R. (2000). Control selection for RNA quantitation. Biotechniques, 29, 332–337.
Mogal, A., & Abdulkadir, S. A. (2006). Effects of histone deacetylase inhibitor (HDACi); Trichostatin-A (TSA) on the expression of housekeeping genes. Molecular and Cellular Probes, 20, 81–86.
Campos, M. S., Rodini, C. O., Pinto-Júnior, D. S., & Nunes, F. D. (2009). GAPD and tubulin are suitable internal controls for qPCR analysis of oral squamous cell carcinoma cell lines. Oral Oncology, 45, 121–126.
Kastl, L., Brown, I., & Schofield, A. C. (2010). Effects of decitabine on the expression of selected endogenous control genes in human breast cancer cells. Molecular and Cellular Probes, 24, 87–92.
Qi, L., & Sit, K. H. (2000). Suicidal differential housekeeping gene activity in apoptosis induced by DCNP. Apoptosis, 5, 379–388.
Barnard, G. F., Staniunas, R. J., Bao, S., Manine, K., Steele, G. D., Jr., Gollan, J. L., et al. (1992). Increased expression of human ribosomal phosphoprotein P0 messenger RNA in hepatocellular carcinoma and colon carcinoma. Cancer Research, 52, 3067–3072.
Henry, J. L., Coggin, D. L., & King, C. R. (1993). High-level expression of the ribosomal protein L19 in human breast tumors that overexpress erbB-2. Cancer Research, 53, 1403–1408.
Vaarala, M. H., Porvari, K. S., Kyllonen, A. P., Mustonen, M. V. J., Lukkarinen, O., & Vihko, P. T. (1998). Several genes encoding ribosomal proteins are over-expressed in prostate-cancer cell lines: Confirmation of L7a and L37 over-expression in prostate-cancer tissue samples. International Journal of Cancer, 78, 27–32.
Haller, F., Kulle, B., Schwager, S., Gunawan, B., von Heydebreck, A., Sültmann, H., et al. (2004). Equivalence test in quantitative reverse transcription polymerase chain reaction: Confirmation of reference genes suitable for normalization. Analytical Biochemistry, 335, 1–9.
Gao, Q., Wang, X.-Y., Fan, J., Qiu, S.-J., Zhou, J., & Shi, Y.-H. (2008). Selection of reference genes for real-time PCR in human hepatocellular carcinoma tissues. Journal of Cancer Research and Clinical Oncology, 134, 979–986.
Fu, L.-Y., Jia, H.-L., Dong, Q.-Z., Wu, J.-C., Zhao, Y., Zhou, H.-J., et al. (2009). Suitable reference genes for real-time PCR in human HBV-related hepatocellular carcinoma with different clinical prognoses. BMC Cancer, 9, 49–60.
Schek, N., Hall, B. L., & Finn, O. J. (1988). Increased glyceraldehyde-3-phosphate dehydrogenase gene expression in human pancreatic adenocarcinoma. Cancer Research, 48, 6354–6359.
Andersen, C. L., Jensen, J. L., & Orntoft, T. F. (2004). Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Research, 64, 5245–5250.
Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., & Kubista, M. (2009). The MIQE Guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clinical Chemistry, 55(4), 611–622.
Bellosillo, B., Piqué, M., Barragán, M., Castaño, E., Villamor, N., & Colomer, D. (1998). Aspirin and salicylate induce apoptosis and activation of caspases in B-cell chronic lymphocytic leukemia cells. Blood, 92(4), 1406–1414.
Piqué, M., Barragán, M., Dalmau, M., Bellosillo, B., Pons, G., & Gil, J. (2000). Aspirin induces apoptosis through mitochondrial cytochrome c release. FEBS Letters, 480, 193–196.
Harris, R. E., Beebe-Donk, J., & Alshafie, G. A. (2006). Reduction in the risk of human breast cancer by selective cyclooxygenase-2 (COX-2) inhibitors. BMC Cancer, 6, 27–31.
Zerbini, L. F., Czibere, A., Wang, Y., Correa, R. G., Out, H., & Joseph, M. (2006). A novel pathway involving melanoma differentiation associated gene-7/interleukin-24 mediates non-steroidal anti-inflammatory drug-induced apoptosis and growth arrest of cancer cells. Cancer Research, 66(24), 11922–11931.
Elder, D. J. E., Hague, A., Hicks, D. J., & Paraskeva, C. (1996). Differential growth inhibition by the aspirin metabolite salicylate in human colorectal tumor cell lines: Enhanced apoptosis in carcinoma and in vitro-transformed adenoma relative to adenoma cell lines. Cancer Research, 56, 2273–2276.
Klampfer, L., Cammenga, J., Wisniewski, H.-G., & Nimer, S. D. (1999). Sodium salicylate activates caspases and induces apoptosis of myeloid leukemia cell lines. Blood, 93(7), 2386–2394.
Sotiriou, C., Lacroix, M., Lagneaux, L., Berchem, G., & Body, J.-J. (1999). The aspirin metabolite salicylate inhibits breast cancer cells growth and their synthesis of the osteolytic cytokines interleukins-6 and -11. Anticancer Research, 19(4B), 2997–3006.
Vandaele, L., Goossens, K., Peelman, L., & Van Soom, A. (2008). mRNA expression of Bcl-2, Bax, caspase-3 and -7 cannot be used as a marker for apoptosis in bovine blastocysts. Animal Reproduction Science, 106, 168–173.
Qi, L., & Sit, K. H. (2000). Housekeeping genes commanded to commit suicide in CpG-cleavage commitment upstream of Bcl-2 inhibition in caspase-dependent and -independent pathways. Molecular Cell Biology Research Communications, 3, 319–327.
Chuang, D.-M., Hough, C., & Senatorov, V. V. (2005). Glyceraldehyde-3-phosphate dehydrogenase, apoptosis and neurodegenerative diseases. Annual Review of Pharmacology and Toxicology, 45, 269–290.
Rho, H.-W., Lee, B.-C., Choi, E.-S., Choi, I.-J., Lee, Y.-S., & Goh, S.-H. (2010). Identification of valid reference genes for gene expression studies of human stomach cancer by reverse transcription-qPCR. BMC Cancer, 10, 240–252.
Almeida, A., Thiery, J. P., Magdelénat, H., & Radvanyi, F. (2004). Gene expression analysis by real-time reverse transcription polymerase chain reaction: Influence of tissue handling. Analytical Biochemistry, 328, 101–108.
Koyama, S., Takashima, Y., Sakurai, T., Suzuki, Y., Taki, M., & Miyakoshi, J. (2007). Effects of 2.45 GHz electromagnetic fields with a wide range of SARs on bacterial and HPRT gene mutations. Journal of Radiation Research, 48(1), 69–75.
Nickells, R. W., & Browder, L. W. (1988). A role for glyceraldehyde-3-phosphate dehydrogenase in the development of thermotolerance in Xenopus laevis embryos. Journal of Cell Biology, 107, 1901–1909.
Goidin, D., Mamessier, A., Staquet, M.-J., Schmitt, D., & Berthier-Vergnes, O. (2001). Ribosomal 18S RNA prevails over glyceraldehyde-3-phosphate dehydrogenase and β-actin genes as internal standard for quantitative comparison of mRNA levels in invasive and non-invasive. Analytical Biochemistry, 295, 17–21.
Aerts, J. L., Gonzales, M. I., & Topalian, S. L. (2004). Selection of appropriate control genes to assess expression of tumor antigens using realtime RT–PCR. Biotechniques, 36, 84–91.
Schimid, H., Cohen, C. D., Henger, A., Irrgang, S., Schlöndorff, D., & Kretzler, M. (2003). Validation of endogenous controls for gene expression analysis in microdissected human renal biopsies. Kidney International, 64, 356–360.
Zhu, L.-J., & Altmann, S. W. (2005). mRNA and 18S-RNA coapplication–reverse transcription for quantitative gene expression analysis. Analytical Biochemistry, 345, 102–109.
Robinson, T. L., Sutherland, I. A., & Sutherland, J. (2007). Validation of candidate bovine reference genes for use with real-time PCR. Veterinary Immunology and Immunopathology, 115, 160–165.
Acknowledgments
This research was supported by the National Research Foundation of South Africa and the Walker Trust Fund of the Faculty of Science, University of Johannesburg, South Africa.
Conflict of interest
The authors of this study have no conflicts of interest or any financial disclosures to make.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ferreira, E., Cronjé, M.J. Selection of Suitable Reference Genes for Quantitative Real-Time PCR in Apoptosis-Induced MCF-7 Breast Cancer Cells. Mol Biotechnol 50, 121–128 (2012). https://doi.org/10.1007/s12033-011-9425-3
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-011-9425-3