Abstract
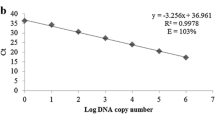
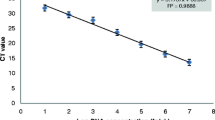
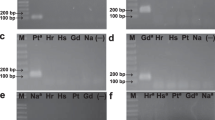
The plant growth promoting bacteria Herbaspirillum seropedicae SmR1 is an endophytic diazotroph found in several economically important crops. Considering that methods to monitor the plant–bacteria interaction are required, our objective was to develop a real-time PCR method for quantification of PGPB H. seropedicae in the rhizosphere of maize seedlings. Primer pairs were designed, and their specificity was verified using DNA from 12 different bacterial species. Ten standard curves of qPCR assay using HERBAS1 primers and tenfold serial dilutions of H. seropedicae SmR1 DNA were performed, and PCR efficiency of 91 % and correlation coefficient of 0.99 were obtained. H. seropedicae SmR1 limit of detection was 101 copies (corresponding to 60.3 fg of bacterial DNA). qPCR assay using HERBAS1 was used to detect and quantify H. seropedicae strain SmR1 in inoculated maize roots, cultivated in vitro and in pots, harvested 1, 4, 7, and 10 days after inoculation. The estimated bacterial DNA copy number per gram of root was in the range 107–109 for plants grown in vitro and it was around 106 for plants grown in pots. Primer pair HERBAS1 was able to quantify H. seropedicae SmR1, and this assay can be useful for monitoring plant–bacteria interaction.





Similar content being viewed by others
References
Andersen, C. B., Holst-Jensen, A., Berdal, K. G., Thorstensen, T., & Tengs, T. (2006). Equal performance of TaqMan, MGB, molecular beacon, and SYBR green-based detection assays in detection and quantification of roundup ready soybean. Journal of Agricultural and Food Chemistry, 54(26), 9658–9663. doi:10.1021/jf061987c.
Bal, H. B., Nayak, L., Das, S., & Adhya, T. K. (2013). Isolation of ACC deaminase producing PGPR from rice rhizosphere and evaluating their plant growth promoting activity under salt stress. Plant and Soil, 366(1–2), 93–105. doi:10.1007/s11104-012-1402-5.
Baldani, J., Baldani, V., Seldin, L., & Dobereiner, J. (1986). Characterization of Herbaspirillum seropedicae Gen-Nov, Sp-Nov, a root-associated nitrogen-fixing bacterium. International Journal of Systematic Bacteriology, 36(1), 86–93.
Baldani, J. I., Pot, B., Kirchhof, G., Falsen, E., Baldani, V. L. D., Olivares, F. L., et al. (1996). Emended description of Herbaspirillum; Inclusion of Pseudomonas rubrisubalbicans, a mild plant pathogen, as Herbaspirillum rubrisubalbicans comb nov; and classification of a group of clinical isolates (EF group 1) as Herbaspirillum species 3. International Journal of Systematic Bacteriology, 46(3), 802–810.
Balsanelli, E., Serrato, R. V., de Baura, V. A., Sassaki, G., Yates, M. G., Rigo, L. U., et al. (2010). Herbaspirillum seropedicae rfbB and rfbC genes are required for maize colonization. Environmental Microbiology, 12(8), 2233–2244. doi:10.1111/j.1462-2920.2010.02187.x.
Bastian, F., Cohen, A., Piccoli, P., Luna, V., Baraldi, R., & Bottini, R. (1998). Production of indole-3-acetic acid and gibberellins A(1) and A(3) by Acetobacter diazotrophicus and Herbaspirillum seropedicae in chemically defined culture media. Plant Growth Regulation, 24(1), 7–11. doi:10.1023/A:1005964031159.
Baudoin, E., Couillerot, O., Spaepen, S., Moenne-Loccoz, Y., & Nazaret, S. (2010). Applicability of the 16S-23S rDNA internal spacer for PCR detection of the phytostimulatory PGPR inoculant Azospirillum lipoferum CRT1 in field soil. Journal of Applied Microbiology, 108(1), 25–38. doi:10.1111/j.1365-2672.2009.04393.x.
Brusamarello-Santos, L. C. C., Pacheco, F., Aljanabi, S. M. M., Monteiro, R. A., Cruz, L. M., Baura, V. A., et al. (2012). Differential gene expression of rice roots inoculated with the diazotroph Herbaspirillum seropedicae. Plant and Soil, 356(1–2), 113–125. doi:10.1007/s11104-011-1044-z.
Cankar, K., Stebih, D., Dreo, T., Zel, J., & Gruden, K. (2006). Critical points of DNA quantification by real-time PCR: Effects of DNA extraction method and sample matrix on quantification of genetically modified organisms. BMC Biotechnology, 6, 37. doi:10.1186/1472-6750-6-37.
Chubatsu, L. S., Monteiro, R. A., de Souza, E. M., Schuler de Oliveira, M. A., Yates, M. G., Wassem, R., et al. (2012). Nitrogen fixation control in Herbaspirillum seropedicae. Plant and Soil, 356(1–2), 197–207. doi:10.1007/s11104-011-0819-6.
Costa, J., Mafra, I., Kuchta, T., & Oliveira, M. B. P. P. (2012). Single-tube nested real-time PCR as a new highly sensitive approach to trace hazelnut. Journal of Agricultural and Food Chemistry, 60(33), 8103–8110. doi:10.1021/jf302898z.
Couillerot, O., Bouffaud, M.-L., Baudoin, E., Muller, D., Caballero-Mellado, J., & Moenne-Loccoz, Y. (2010). Development of a real-time PCR method to quantify the PGPR strain Azospirillum lipoferum CRT1 on maize seedlings. Soil Biology & Biochemistry, 42(12), 2298–2305. doi:10.1016/j.soilbio.2010.09.003.
de Souza, R., Beneduzi, A., Ambrosini, A., da Costa, P. B., Meyer, J., Vargas, L. K., et al. (2013). The effect of plant growth-promoting rhizobacteria on the growth of rice (Oryza sativa L.) cropped in southern Brazilian fields. Plant and Soil, 366(1–2), 585–603. doi:10.1007/s11104-012-1430-1.
Dinon, A., Prins, T., van Dijk, J., Arisi, A., Scholtens, I., & Kok, E. (2011). Development and validation of real-time PCR screening methods for detection of cry1A.105 and cry2Ab2 genes in genetically modified organisms. Analytical and Bioanalytical Chemistry, 400(5), 1433–1442. doi:10.1007/s00216-011-4875-9.
Egener, T., Hurek, T., & Reinhold-Hurek, B. (1999). Endophytic expression of nif genes of Azoarcus sp strain BH72 in rice roots. Molecular Plant–Microbe Interactions, 12(9), 813–819. doi:10.1094/mpmi.1999.12.9.813.
Elbeltagy, A., Nishioka, K., Sato, T., Suzuki, H., Ye, B., Hamada, T., et al. (2001). Endophytic colonization and in planta nitrogen fixation by a Herbaspirillum sp. isolated from wild rice species. Applied and Environment Microbiology, 67(11), 5285–5293. doi:10.1128/AEM.67.11.5285-. 5293.2001.
Faleiro, A., Pereira, T., Espindula, E., Brod, F., & Arisi, A. (2013). Real time PCR detection targeting nifA gene of plant growth promoting bacteria Azospirillum brasilense strain FP2 in maize roots. Symbiosis, 61(3), 125–133. doi:10.1007/s13199-013-0262-y.
Fan, B., Chen, X. H., Budiharjo, A., Bleiss, W., Vater, J., & Borriss, R. (2011). Efficient colonization of plant roots by the plant growth promoting bacterium Bacillus amyloliquefaciens FZB42, engineered to express green fluorescent protein. Journal of Biotechnology, 151(4), 303–311. doi:10.1016/j.jbiotec.2010.12.022.
Gaut, B. S., d’Ennequin, M. L., Peek, A. S., & Sawkins, M. C. (2000). Maize as a model for the evolution of plant nuclear genomes. Proceedings of the National Academy of Sciences of the United States of America, 97(13), 7008–7015. doi:10.1073/pnas.97.13.7008.
Gyaneshwar, P., James, E. K., Reddy, P. M., & Ladha, J. K. (2002). Herbaspirillum colonization increases growth and nitrogen accumulation in aluminium-tolerant rice varieties. New Phytologist, 154(1), 131–145. doi:10.1046/j.1469-8137.2002.00371.x.
James, E. K., Gyaneshwar, P., Mathan, N., Barraquio, Q. L., Reddy, P. M., Iannetta, P. P. M., et al. (2002). Infection and colonization of rice seedlings by the plant growth-promoting bacterium Herbaspirillum seropedicae Z67. Molecular Plant–Microbe Interactions, 15(9), 894–906. doi:10.1094/mpmi.2002.15.9.894.
James, E. K., & Olivares, F. L. (1998). Infection and colonization of sugar cane and other graminaceous plants by endophytic diazotrophs. Critical Reviews in Plant Sciences, 17(1), 77–119. doi:10.1016/s0735-2689(98)00357-8.
James, E. K., Olivares, F. L., Baldani, J. I., & Dobereiner, J. (1997). Herbaspirillum, an endophytic diazotroph colonizing vascular tissue in leaves of Sorghum bicolor L Moench. Journal of Experimental Botany, 48(308), 785–797. doi:10.1093/jxb/48.3.785.
Kang, M., Kim, M., Hwang, D., Cho, M., Seol, Y., Hahn, J., et al. (2012). Quantitative in planta PCR assay for specific detection of Xanthomonas oryzae pv. oryzicola using putative membrane protein based primer set. Crop Protection, 40, 22–27. doi:10.1016/j.cropro.2012.04.014.
Klassen, G., Pedrosa, F. O., Souza, E. M., Funayama, S., & Rigo, L. U. (1997). Effect of nitrogen compounds on nitrogenase activity in Herbaspirillum seropedicae SMR1. Canadian Journal of Microbiology, 43(9), 887–891.
Liu, F. C., Xing, S. J., Ma, H. L., Du, Z. Y., & Ma, B. Y. (2013). Plant growth-promoting rhizobacteria affect the growth and nutrient uptake of Fraxinus americana container seedlings. Applied Microbiology and Biotechnology, 97(10), 4617–4625. doi:10.1007/s00253-012-4255-1.
Monteiro, R. A., Balsanelli, E., Tuleski, T., Faoro, H., Cruz, L. M., Wassem, R., et al. (2012). Genomic comparison of the endophyte Herbaspirillum seropedicae SmR1 and the phytopathogen Herbaspirillum rubrisubalbicans M1 by suppressive subtractive hybridization and partial genome sequencing. FEMS Microbiology Ecology, 80(2), 441–451. doi:10.1111/j.1574-6941.2012.01309.x.
Monteiro, R. A., Balsanelli, E., Wassem, R., Marin, A. M., Brusamarello-Santos, L. C. C., Schmidt, M. A., et al. (2012). Herbaspirillum-plant interactions: microscopical, histological and molecular aspects. Plant and Soil, 356(1–2), 175–196. doi:10.1007/s11104-012-1125-7.
Monteiro, R. A., Schmidt, M. A., de Baura, V. A., Balsanelli, E., Wassem, R., Yates, M. G., et al. (2008). Early colonization pattern of maize (Zea mays L. Poales, Poaceae) roots by Herbaspirillum seropedicae (Burkholderiales, Oxalobacteraceae). Genetics and Molecular Biology, 31(4), 932–937. doi:10.1590/s1415-47572008005000007.
Pedrosa, F. O., Monteiro, R. A., Wassem, R., Cruz, L. M., Ayub, R. A., Colauto, N. B., et al. (2011). Genome of Herbaspirillum seropedicae strain SmR1, a specialized diazotrophic endophyte of tropical grasses. Plos Genetics, 7(5), e1002064. doi:10.1371/journal.pgen.1002064.
Raj, M., Jeeva, M. L., Hegde, V., Vidyadharan, P., Archana, P. V., Senthil alias Sankar, M., et al. (2012). Polymerase chain reaction assay for rapid, sensitive detection, and identification of Colletotrichum gloeosporioides causing greater yam anthracnose. Molecular Biotechnology, 52(3), 277–284. doi:10.1007/s12033-012-9496-9.
Rashid, S., Charles, T. C., & Glick, B. R. (2012). Isolation and characterization of new plant growth-promoting bacterial endophytes. Applied Soil Ecology, 61, 217–224. doi:10.1016/j.apsoi1.2011.09.011.
Rodriguez-Salazar, J., Suarez, R., Caballero-Mellado, J., & Iturriaga, G. (2009). Trehalose accumulation in Azospirillum brasilense improves drought tolerance and biomass in maize plants. FEMS Microbiology Letters, 296(1), 52–59. doi:10.1111/j.1574-6968.2009.01614.x.
Roncato-Maccari, L. D. B., Ramos, H. J. O., Pedrosa, F. O., Alquini, Y., Chubatsu, L. S., Yates, M. G., et al. (2003). Endophytic Herbaspirillum seropedicae expresses nif genes in gramineous plants. FEMS Microbiology Ecology, 45(1), 39–47. doi:10.1016/s0168-6496(03)00108-9.
Roncato-Maccari, L. D. B., Ramos, H. J. O., Pedrosa, F. O., Alquini, Y., Chubatsu, L. S., Yates, M. G., et al. (2003). Root colonization, systemic spreading and contribution of Herbaspirillum seropedicae to growth of rice seedling. Symbiosis, 35(1–3), 261–270.
Ruppel, S., Ruhlmann, J., & Merbach, W. (2006). Quantification and localization of bacteria in plant tissues using quantitative real-time PCR and online emission fingerprinting. Plant and Soil, 286(1–2), 21–35. doi:10.1007/s11104-006-9023-5.
Schmidt, M., Souza, E., Baura, V., Wassem, R., Yates, M., Pedrosa, F., et al. (2011). Evidence for the endophytic colonization of Phaseolus vulgaris (common bean) roots by the diazotroph Herbaspirillum seropedicae. Brazilian Journal of Medical and Biological Research, 44(3), 182–185. doi:10.1590/S0100-879X2011007500004.
Schnable, P. S. (2012). The B73 maize genome: Complexity, diversity, and dynamics (November, pg 1112, 2009). Science, 337(6098), 1040.
Shime-Hattori, A., Kobayashi, S., Ikeda, S., Asano, R., Shime, H., & Shinano, T. (2011). A rapid and simple PCR method for identifying isolates of the genus Azospirillum within populations of rhizosphere bacteria. Journal of Applied Microbiology, 111(4), 915–924. doi:10.1111/j.1365-2672.2011.05115.x.
Stets, M. I., Pinto, A. S, Jr, Huergo, L. F., de Souza, E. M., Guimaraes, V. F., Alves, A. C., et al. (2013). Rapid identification of bacterial isolates from wheat roots by high resolution whole cell MALDI-TOF MS analysis. Journal of Biotechnology, 165(3–4), 167–174. doi:10.1016/j.jbiotec.2013.04.001.
Su’udi, M., Kim, J., Park, J.-M., Bae, S.-C., Kim, D., Kim, Y.-H., et al. (2013). Quantification of rice blast disease progressions through Taqman real-time PCR. Molecular Biotechnology, 55(1), 43–48. doi:10.1007/s12033-012-9632-6.
Taule, C., Mareque, C., Barlocco, C., Hackembruch, F., Reis, V. M., Sicardi, M., et al. (2012). The contribution of nitrogen fixation to sugarcane (Saccharum officinarum L.), and the identification and characterization of part of the associated diazotrophic bacterial community. Plant and Soil, 356(1–2), 35–49. doi:10.1007/s11104-011-1023-4.
Timmusk, S., Paalme, V., Lagercrantz, U., & Nevo, E. (2009). Detection and quantification of Paenibacillus polymyxa in the rhizosphere of wild barley (Hordeum spontaneum) with real-time PCR. Journal of Applied Microbiology, 107(3), 736–745. doi:10.1111/j.1365-2672.2009.04265.x.
Verma, S. C., Ladha, J. K., & Tripathi, A. K. (2001). Evaluation of plant growth promoting and colonization ability of endophytic diazotrophs from deep water rice. Journal of Biotechnology, 91(2–3), 127–141. doi:10.1016/s0168-1656(01)00333-9.
Videira, S. S., de Oliveira, D. M., de Morais, R. F., Borges, W. L., Divan Baldani, V. L., & Baldani, J. I. (2012). Genetic diversity and plant growth promoting traits of diazotrophic bacteria isolated from two Pennisetum purpureum Schum. genotypes grown in the field. Plant and Soil, 356(1–2), 51–66. doi:10.1007/s11104-011-1082-6.
Zhang, T., & Fang, H. H. (2006). Applications of real-time polymerase chain reaction for quantification of microorganisms in environmental samples. Applied Microbiology and Biotechnology, 70(3), 281–289. doi:10.1007/s00253-006-0333-6.
Acknowledgments
We would like to express our gratitude to Fábio de Oliveira Pedrosa, Leda Chubatsu, and Michelle Tadra-Sfeir, Universidade Federal do Paraná, for providing A. brasilense strain FP2, H. seropedicae strain SmR1, and other Herbaspirillum, to Luciane Passaglia for providing Rhizobium, Microbacterium, and Pseudomonas. This work was financially supported by the National Institute of Science and Technology—Biological Nitrogen Fixation, INCT-FBN, CNPq, Ministry of Science and Technology, Brazil. TPP, FPA, and PD were recipients of Master and PhD fellowships from CAPES, Ministry of Education, Brazil. FCAB is recipient of post-doctoral fellowship from CAPES PNPD, and ACMA is recipient of research fellowship from CNPq (PQ-2).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Pereira, T.P., do Amaral, F.P., Dall’Asta, P. et al. Real-Time PCR Quantification of the Plant Growth Promoting Bacteria Herbaspirillum seropedicae Strain SmR1 in Maize Roots. Mol Biotechnol 56, 660–670 (2014). https://doi.org/10.1007/s12033-014-9742-4
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12033-014-9742-4