Abstract
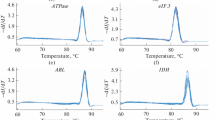
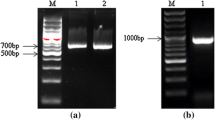
Quantitative real-time reverse transcription-polymerase chain reaction (qRT-PCR) is a well-known method to quantify gene expression by comparing with the reference genes. Generally, housekeeping genes were set as references, as for their stable expression in varying conditions. Here, we try to evaluate few of such genes to identify suitable housekeeping genes as references for qRT-PCR analysis of gene expression in Misgurnus anguillicaudatus. This study evaluated the expression of four commonly used housekeeping genes, i.e. b-actin (ACTB), elongation factor 1 alpha (EF-1a), glyceraldehyde-3-phosphate (GAPDH) and 18S ribosomal RNA (18S rRNA), in gender difference, effects of tissue type, different developmental stages, chemical treatment of embryos/larvae with commonly used vehicles for administration and agents that represent known environmental toxicant. Rank ordering of expression stability was done using geNorm, NormFinder and BestKeeper algorithms. Results suggested that in the qRT-PCR test, in all the experimental conditions, EF-1a could be selected as reference gene when analysing a target gene. For the study of different development stages, ACTB could be a candidate as reference gene. For the studies associated with different gender and tissue types, EF-1a would be better targeted as reference gene. Meanwhile, in toxicant treatment, expression of EF-1a seems to be more stable than others and could be considered as reference gene. This study could provide useful guidelines that can be expected to aid M. anguillicaudatus researchers in their initial choice of housekeeping genes for future studies and enable more accurate normalization of gene expression data.





Similar content being viewed by others
References
Andersen C. L., Jensen J. L. and Orntoft T. F. 2004 Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64, 5245–5250.
ASAB 2003 Guidelines for the treatment of animals in behavioural research and teaching. Anim. Behav. 65, 249–255.
Bower N. I. and Johnston I. A. 2009 Selection of reference genes for expression studies with fish myogenic cell cultures. BMC Mol. Biol. 10, 80.
Bustin S. A. 2000 Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J. Mol. Endocrinol. 25, 169–193.
Caradec J., Sirab N., Keumeugni C., Moutereau S., Chimingqi M., Matar C. et al. 2010 Desperate house genes: the dramatic example of hypoxia. Br. J. Cancer 102, 1037–1043.
Chang E., Shi S., Liu J., Cheng T., Xue L., Yang X. et al. 2012 Selection of reference genes for quantitative gene expression studies in Platycladus orientalis (Cupressaceae) using real-time PCR. PLoS One 7, e33278.
Cruz F., Kalaoun S., Nobile P., Colombo C., Almeida J., Barros L. M. G. et al. 2009 Evaluation of coffee reference genes for relative expression studies by quantitative real-time RT-PCR. Mol. Breed. 23, 607–616.
Dheda K., Huggett J. F., Bustin S. A., Johnson M. A., Rook G. and Zumla A. 2004 Validation of housekeeping genes for normalizing RNA expression in real-time PCR. Biotechniques 37, 112–119.
Dheda K., Huggett J. F., Chang J. S., Kim L. U., Bustin S. A., Johnson M. A. et al. 2005 The implications of using an inappropriate reference gene for real-time reverse transcription PCR data normalization. Anal. Biochem. 344, 141–143.
Fernandez J. M. O., Mommens M., Hagen Ø., Babiak I. and Solberg C. 2008 Selection of suitable reference genes for real-time PCR studies of Atlantic halibut development. Comp. Biochem. Physiol. B, Biochem. Mol. Biol. 150, 23–32.
Filby A. L. and Tyler C. R. 2007 Appropriate ‘housekeeping’ genes for use in expression profiling the effects of environmental estrogens in fish. BMC Mol. Biol. 8, 10.
Frost P. and Nilsen F. 2003 Validation of reference genes for transcription profiling in the salmon louse, Lepeophtheirus salmonis, by quantitative real-time PCR. Vet. Parasitol. 118, 169–174.
Gilsbach R., Kouta M., Boenisch H. and Bruess M. 2006 Comparison of in vitro and in vivo reference genes for internal standardization of real-time PCR data. BioTechniques 40, 173–177.
Glare E. M., Divjak M., Bailey M. J. and Walters E. H. 2002 Beta-actin and GAPDH housekeeping gene expression in asthmatic airways is variable and not suitable for normalising mRNA levels. Thorax 57, 765–770.
Hendriks-Balk M. C., Michel M. C. and Alewijnse A. E. 2007 Pitfalls in the normalization of real-time polymerase chain reaction data. Basic Res. Cardiol. 102, 195–197.
Hildahl J., Sweeney G., Galay-Burgos M., Einarsdóttir I. E. and Björnsson B. T. 2007 Cloning of Atlantic halibut growth hormone receptor genes and quantitative gene expression during metamorphosis. Gen. Comp. Endocrinol. 151, 143–152.
Huggett J., Dheda K., Bustin S. and Zumla A. 2005 Real-time qRT-PCR normalisation; strategies and considerations. Genes Immun. 6, 279–284.
Infante C., Matsuoka M. P., Asensio E., Canavate J. P., Reith M. and Manchado M. 2008 Selection of housekeeping genes for gene expression studies in larvae from flatfish using real-time PCR. BMC Mol. Biol. 9, 28.
Jørgensen S. M., Kleveland E. J., Grimholt U. and Gjøen T. 2006 Validation of reference genes for real-time polymerase chain reaction studies in Atlantic salmon. Mar. Biotechnol. 8, 398–408.
Long X. Y., Wang J. R., Ouellet T., Rocheleau H., Wei Y. M., Pu Z. E. et al. 2010 Genome-wide identification and evaluation of novel internal control genes for Q-PCR based transcript normalization in wheat. Plant Mol. Biol. 74, 307–311.
Ma L., Wang W., Liu C., Yu H., Wang Z., Wang X. et al. 2013 Selection of reference genes for reverse transcription quantitative real-time PCR normalization in black rockfish (Sebastes schlegeli). Mar. Genomics 11, 67–73.
Mascia T., Santovito E., Gallitelli D. and Cillo F. 2010 Evaluation of reference genes for quantitative reverse-transcription polymerase chain reaction normalization in infected tomato plants. Mol. Plant Pathol. 11, 805–816.
McCurley A. T. and Callard G. V. 2008 Characterization of housekeeping genes in zebrafish: male-female differences and effects of tissue type, developmental stage and chemical treatment. BMC Mol. Biol. 9, 102.
Mishra R. K. and Eliceiri G. L. 1997 Three small nucleolar RNAs that are involved in ribosomal RNA precursor processing. Proc. Natl. Acad. Sci. USA 94, 4972–4977.
Morga B., Arzul I., Faury N. and Renault T. 2010 Identification of genes from flat oyster Ostrea edulis as suitable housekeeping genes for quantitative real time PCR. Fish Shellfish Immunol. 29, 937–945.
Murphy C. L. and Polak J. M. 2002 Differentiating embryonic stem cells: GAPDH, but neither HPRT nor beta-tuba1 is suitable as an internal standard for measuring RNA levels. Tissue Eng. 8, 551–559.
Nygard A. B., Jorgenson C. B., Cierra S. and Fredholm M. 2007 Selection of reference genes for gene expression studies in pig tissues using SYBR green qRT-PCR. BMC Mol. Biol. 8, 67.
Oliveriera J. G., Prados R. Z., Guedes A. C., Ferreira P. C. and Kroon E. G. 1999 The housekeeping gene glyceraldehyde-3-phosphate dehydrogenase is inappropriate as internal control in comparative studies between skin tissue and cultured skin fibroblasts using Northern blot analysis. Arch. Dermatol. Res. 291, 659–661.
Olsvik P. A., Lie K. K., Jordal A. E., Nilsen T. O. and Hordvik I. 2005 Evaluation of potential reference genes in real-time RT-PCR studies of Atlantic salmon. BMC Mol. Biol. 6, 21.
Olsvik P. A., Søfteland L. and Lie K. K. 2008 Selection of reference genes for qRT-PCR examination of wild populations of Atlantic cod Gadus morhua. BMC Res. Notes 1, 47.
Paria A., Dong J., Suresh Babu P. P., Makesh M., Chaudhari A., Thirunavukkarasu A. R. et al. 2016 Evaluation of candidate reference genes for quantitative expression studies in Asian seabass (Lates calcarifer) during ontogenesis and in tissues of healthy and infected fishes. Indian J. Exp. Biol. 54, 597–605.
Pfaffl M. W., Tichopad A., Prgomet C. and Neuvians T. P. 2004 Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: Best keeper–excel-based tool using pair-wise correlations. Biotechnol. Lett. 26, 509–515.
Robledo D., Jorge H. U., Rosa M. C., Belén G. P., Laura S., Paulino M. and Ana V. 2014 Analysis of qPCR reference gene stability determination methods and a practical approach for efficiency calculation on a turbot (Scophthalmus maximus) gonad dataset. BMC Genomics 15, 648.
Radonic A., Thulke S., Mackay I. M., Landt O., Siegert W. and Nitsche A. 2004 Guideline to reference gene selection for quantitative real-time PCR. Biochem. Biophys. Res. Commun. 313, 856–862.
Schmittgen T. D. and Zakrajsek B. A. 2000 Effect of experimental treatment on housekeeping gene expression: validation by real-time, quantitative RT-PCR. J. Biochem. Biophys. Methods 46, 69–81.
Selvey S., Thompson E. W., Matthaei K., Lea R. A., Irving M. G. and Griffiths L. R. 2001 Beta actin–an unsuitable internal control for RT-PCR. Mol. Cell. Probes 15, 307–311.
Silver N., Best S., Jiang J. and Thein S. 2006 Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 7, 33.
Solanas M., Moral R. and Escrich E. 2001 Unsuitability of using ribosomal RNA as loading control for Northern blot analyses related to the imbalance between messenger and ribosomal RNA content in rat mammary tumors. Anal. Biochem. 288, 99–102.
Tang R., Dodd A., Lai D., McNabb W. C. and Love D. R. 2007 Validation of zebrafish (Danio rerio) reference genes for quantitative real-time RT-PCR normalization. Acta Biochim Biophys. Sin. 39, 384–390.
Thellin O., Zorzi W., Lakaye B., De Borman B., Coumans B., Hennen G. et al. 1999 Housekeeping genes as internal standards: use and limits. J. Biotechnol. 75, 291–295.
Tricarico C., Pinzani P., Bianchi S., Paglierani M., Distante V., Pizzagli M. et al. 2002 Quantitative real-time reverse transcription polymerase chain reaction: normalization to rRNA or single housekeeping genes is inappropriate for human tissue biopsies. Anal. Biochem. 309, 293–300.
Urbatzka R., Galante-Oliveira S., Rocha E., Castro L. F. C. and Cunha I. 2013 Normalization strategies for gene expression studies by real-time PCR in a marine fish species, Scophthalmus maximus. Mar. Genomics 10, 17.
Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A. and Speleman F. 2002 Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, research0034.1–research0034.11.
Zhang Z. and Hu J. 2007 Development and validation of endogenous reference genes for expression profiling of medaka (Oryzias latipes) exposed to endocrine disrupting chemicals by quantitative real-time RT-PCR. Toxicol. Sci. 95, 356–368.
Zhong H. and Simons J. W. 1999 Direct comparison of GAPDH, betaactin, cyclophilin, and 28S rRNA as internal standards for quantifying RNA levels under hypoxia. Biochem. Biophys. Res. Commun. 259, 523–526.
Zhong Q., Zhang Q., Wang Z., Qi J., Chen Y., Li S. et al. 2008 Expression profiling and validation of potential reference genes during paralichthys olivaceus embryogenesis. Mar. Biotechnol. 3, 310–318.
Acknowledgements
This work is supported by grants from the National Natural Science Foundation of China (no. 31200923), Young Core Instructor Foundation of Henan Normal University (no. 5101049470610) and Doctor Subject Foundation of Henan Normal University (no. 01046500109).
Author information
Authors and Affiliations
Corresponding author
Additional information
Corresponding editor: Rajiva Raman
Rights and permissions
About this article
Cite this article
Xia, X., Huo, W., Wan, R. et al. Identification of housekeeping genes as references for quantitative real-time RT-PCR analysis in Misgurnus anguillicaudatus . J Genet 96, 895–904 (2017). https://doi.org/10.1007/s12041-017-0845-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12041-017-0845-0