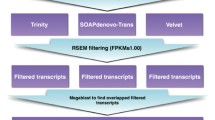
Abstract
The Chinese forest musk deer (Moschus berezovskii Flerov) is an endangered artiodactyl mammal. The musk secreted by sexually mature males is highly valued for alleged pharmaceutical properties and perfume manufacturing. However, the genomic and transcriptomic resources of musk deer remain deficiently represented and poorly understood. Next-generation sequencing technique is an efficient method for generating an enormous amount of sequence data that can represent a large number of genes and their expression levels. In the present study, we used Illumina HiSeq technology to perform de novo assembly of heart and musk gland transcriptomes from the Chinese forest musk deer. A total of 239,383 transcripts and 176,450 unigenes were obtained, of which 37,329 unigenes were matched to known sequences in the NCBI nonredundant protein (Nr) database; 31,039 unigenes were assigned to 61 GO terms, and 11,782 to 332 KEGG pathways. Additionally, 592 and 2282 differentially expressed genes were found to be specifically expressed in the heart and musk gland, respectively. The abundant transcriptomic data generated in the present report will provide a comprehensive sequence resource for Chinese forest musk deer as well as lay down a foundation which will help in accelerating genetic and functional genomics research in this species.



Similar content being viewed by others
References
Anders S. and Huber W. 2013 Differential expression of RNA-seq data at the gene level-the DESeq package (available at www.bioconductor.org/packages/devel/bioc/vignettes/DESeq/inst/doc/DESeq.pdf).
Audic S. and Claverie J. M. 1997 The significance of digital gene expression profiles. Genome Res. 7, 986–995.
Bi S. Z. and Shen Y. 1986 Study on morphology and chemical communication mechanism in musk gland. Chinese J. Zool. 02, 11–14.
Bi S. Z., Shen Y., Zhu D. X., Zhu C. S., Jia L. Z. and Zhang Z. M. 1984 Study on ultra-microstructure and musk secretion of musk gland in prosperous secreting stage. Acta Ther. Sinica 02, 81–85.
Caillier B., Lépine J., Tojcic J., Ménard V., Perusse L., Bélanger A. et al. 2007 A pharmacogenomics study of the human estrogen glucuronosyltransferase UGT1A3. Pharmacogenet Gen. 17, 481–495.
Cao X. H. and Zhou Y. D. 2007 Progress on anti-inflammatory effects of musk. China pharm. 18, 1662–1665.
Chen H. Y., Shen H., Jia B., Zhang Y. S., Wang X. H. and Zeng X. C. 2015 Differential gene expression in ovaries of Qira black sheep and Hetian sheep using RNA-Seq technique. PLoS One 10, e0120170.
Chen X. 2007 Studies on the genetic diversity of forest musk deer (Moschus berezovskii) and linkage analysis between the performance of musk productivity and AFLP markers. MD thesis, Zhejiang University, Hangzhou.
Conesa A., Götz S., Garcíagómez J. M., Terol J., Talón M. and Robles M. 2005 Blast2go: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674.
Crowley R. K., Hughes B., Gray J., McCarthy T., Hughes S., Shackleton C. H. et al. 2014 Longitudinal changes in glucocorticoid metabolism are associated with later development of adverse metabolic phenotype. Eur. J. Endocrinol. 171, 433–442.
Deng F. M. 1986 Domestication and grazing control of forest musk deer (Moschus berezovskii). Chinese J. Wildlife 4, 35–37.
Deng T., Pang C., Lu X., Zhu P., Duan A. and Tan Z. 2016 De novo transcriptome assembly of the Chinese swamp buffalo by RNA sequencing and SSR marker discovery. PLoS One 11, e0147132.
Feng Q. Q. and Liu T. J. 2015 Progress on pharmacological activity of muscone. Food Drug 3, 212–214.
Grabherr M. G., Haas B. J., Moran Y., Levin J. Z., Thompson D. A. and Ido A. 2011 Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652.
Guan T. L., Zeng B., Peng Q. K., Yue B. S. and Zou F. D. 2009 Microsatellite analysis of the genetic structure of captive forest musk deer populations and its implication for conservation. Biochem. Syst. Ecol. 37, 166–173.
Haas B. J., Papanicolaou A., Yassour M., Grabherr M., Blood P. D. and Bowden J. 2013 De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512.
Harris M. A., Clark J. and Ireland A. 2004 The gene ontology (GO) database and informatics resource. Nucleic Acids Res. 32, 258–261.
Jia B. Y., Ba H. X., Wang G. W., Yang Y., Cui X. Z. et al. 2016 Transcriptome analysis of sika deer in China. Mol. Genet. Genomics 291, 1–13.
Jiang M., Klein M., Zanger U. M., Mohammad M. K., Cave M. C., Gaikwad N. W. et al. 2016 Inflammatory regulation of steroid sulfatase: A novel mechanism to control estrogen homeostasis and inflammation in chronic liver disease. J. Hepatol. 64, 44–52.
Kanehisa M. 2008 The KEGG database. In In silico simulation of biological processes: novartis foundation symposium 247. (ed. G. Bock and J. A. Gorde). John Wiley, Chichester (https://doi.org/10.1002/0470857897.ch8).
Li B. and Dewey C. N. 2011 Rsem: accurate transcript quantification from rna-seq data with or without a reference genome. BMC Bioinformatics 12, 93–99.
Li D. Y., Chen B. L., Zhang L., Gaur U., Ma T. Y. and Jie H. 2016 The musk chemical composition and microbiota of Chinese forest musk deer males. Sci. Rep. 6, 18975.
Liu M., Yao B., Zhang H., Guo H., Hu D. and Wang Q. 2014 Identification of novel reference genes using sika deer antler transcriptome expression data and their validation for quantitative gene expression analysis. Genes Genom. 36, 573–582.
Martin M. 2011 Cutadapt removes adapter sequences from high-throughput sequencing reads. Embnet J. 17, 10–12.
Martin J. A. and Wang Z. 2011 Next-generation transcriptome assembly. Nat. Rev. Genet. 12, 671–682.
Mortazavi A., Williams B. A., Mccue K., Schaeffer L. and Wold B. 2008 Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 5, 621–628.
Moriya Y., Itoh M., Okuda S., Yoshizawa A. C. and Kanehisa M. 2007 Kaas: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35, 182–185.
Ockendon N. F., O’Connell L. A., Bush S. J., Monzón-Sandoval J., Barnes H. and Székely T. 2015 Optimization of next-generation sequencing transcriptome annotation for species lacking sequenced genomes. Mol. Ecol. Resour. 16, 446–458.
Peng H., Liu S., Zou F., Zeng B. and Yue B. 2009 Genetic diversity of captive forest musk deer (Moschus berezovskii) inferred from the mitochondrial DNA control region. Anim. Genet. 40, 65–72.
Schmieder R. and Edwards R. 2011 Quality control and preprocessing of metagenomic datasets. Bioinforma Oxf. Engl. 27, 863–864.
Seth S. D., Mukhopadhyay A. B. and Prbhakar M. C. 1973 Antihista–minic and spasmolytic effects of musk. JPN. J. Pharmacol. 23, 673–679.
Sheng H. L. 1996 Protection and utilization of musk deer resources in China. Chinese J. Wildlife 91, 10–12.
Strickler S. R., Aureliano B. and Mueller L. A. 2012 Designing a transcriptome next-generation sequencing project for a nonmodel plant species. Am. J. Bot. 99, 257–266.
Su B., Wang Y. X. and Wang Q. S. 2001 Mitochondrial DNA sequences imply Anhui musk deer a valid species in genus Moschus. Zool. Res. 22, 169–173.
Wang X., Gérard C., Thériault J. F., Poirier D., Doillon C. J. and Lin S. X. 2015 Synergistic control of sex hormones by 17\(\upbeta \)–HSD type 7: a novel target for estrogen-dependent breast cancer. J. Mol. Cell Biol. 7, 568–579.
Wang Y. and Harris R. 2015 Moschus berezovskii. (errata version published in 2016) The IUCN Red List of Threatened Species 2015: e.T13894A103431781. (downloaded on 09 January 2017).
Wang Z., Li M., Li L., Sun H. and Lin X. Y. 2015 Association of single nucleotide polymorphisms in the CYP1B1 gene with the risk of primary open-angle glaucoma: a meta-analysis. Genet. Mol. Res. 14, 17262–17272.
Xie C., Mao X., Huang J., Ding Y., Wu J., Dong S. et al. 2011 KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 39, 316–322.
Young M. D., Wakeeld M. J., Smyth G. K. and Oshlack A. 2012 goseq: Gene ontology testing for RNA-seq datasets. 1–25.
Zhang Z., Deng Z. and Li Z. C. 1985 Domestication and trans-cultivation of forest musk deer (Moschus berezovskii). J. Chinese Med. Mater. 2, 14–15.
Zhao S. S. 2009 Assement of genetic diversity in the captive forest musk deer (Moschus berezovskii) and linkage analysis between the performance of musk productivity and DNA molecular markers. Ph.D. thesis, Zhejiang University, Hangzhou.
Zhao Y., Yao B., Zhang M., Wang S., Zhang H. and Xiao W. 2013 Comparative analysis of differentially expressed genes in sika deer antler at different stages. Mol. Biol. Rep. 40, 1665–1676.
Acknowledgements
This study was supported by National Natural Science Foundation of China (NSFC31672396 and NSFC81503188); the programme from Sichuan Agricultural University (02920400) and Sichuan Provincial Department of Science and Technology Programme (2015JQO023).
Author information
Authors and Affiliations
Corresponding author
Additional information
Corresponding editor: Indrajit Nanda
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xu, Z., Jie, H., Chen, B. et al. Illumina-based de novo transcriptome sequencing and analysis of Chinese forest musk deer. J Genet 96, 1033–1040 (2017). https://doi.org/10.1007/s12041-017-0872-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12041-017-0872-x