Abstract
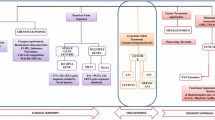
Classification of microorganisms on the basis of traditional microbiological methods (morphological, physiological and biochemical) creates a blurred image about their taxonomic status and thus needs further clarification. It should be based on a more pragmatic approach of deploying a number of methods for the complete characterization of microbes. Hence, the methods now employed for bacterial systematics include, the complete 16S rRNA gene sequencing and its comparative analysis by phylogenetic trees, DNA-DNA hybridization studies with related organisms, analyses of molecular markers and signature pattern(s), biochemical assays, physiological and morphological tests. Collectively these genotypic, chemotaxonomic and phenotypic methods for determining taxonomic position of microbes constitute what is known as the ‘polyphasic approach’ for bacterial systematics. This approach is currently the most popular choice for classifying bacteria and several microbes, which were previously placed under invalid taxa have now been resolved into new genera and species. This has been possible owing to rapid development in molecular biological techniques, automation of DNA sequencing coupled with advances in bioinformatic tools and access to sequence databases. Several DNA-based typing methods are known; these provide information for delineating bacteria into different genera and species and have the potential to resolve differences among the strains of a species. Therefore, newly isolated strains must be classified on the basis of the polyphasic approach. Also previously classified organisms, as and when required, can be reclassified on this ground in order to obtain information about their accurate position in the microbial world. Thus, current techniques enable microbiologists to decipher the natural phylogenetic relationships between microbes.
Similar content being viewed by others
References
Gevers D, Cohan FM, Lawrence JG, Spratt BG, Coenye T, Feil EJ, Stackebrandt E, Van de Peer Y, Vandamme P, Thompson FL & Swings J (2005) Opinion: Re-evaluating prokaryotic species Nat Rev Microbiol 3:733–739
Coenye T, Gevers D, Van de Peer Y, Vandamme P & Swings J (2005) Towards a prokaryotic genomic taxonomy. FEMS Microbiol Rev 29:147–167
Mora RR & Amann R (2001) The species concept for prokaryotes FEMS Microbiol Rev 25:39–67
Vandamme P, Pot B, Gillis M, De Vos P, Kersters K, Swings J (1996) Polyphasic taxonomy, a consensus approach to bacterial systematics Microbiol Rev 60:407–438
Amann RI, Ludwig W & Schleifer KH (1995) Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 59: 143–169
Mayr E and Ashlock PD (1991) Principles of Systematic Zoology 2nd ed. McGraw-Hill, Inc. pp 1–12
Simpson GG (1961) Principles of Animal Taxonomy New York: Columbia University Press
Clarridge JE (2004) Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases Clin Microbiol Rev 17:840–862
Woese CR (1987) Bacterial evolution Microbiol Rev 51:221–272
Schildkraut CL, Marmur J & Doty P (1961) The formation of hybrid DNA molecules and their use in studies of DNA homologies J Mol Biol 3:595–617
Colwell RR (1970) Polyphasic taxonomy of the genus Vibrio: numerical taxonomy of Vibrio cholerae, Vibrio parahaemolyticus and related Vibrio species J Bacteriol 104:410–433
Mayr E (1942) Systematics and the origin of species New York: Columbia University Press
Grimont F & Grimont PAD (1991) DNA fingerprinting In: Stackebrandt E & Goodfellow M Nucleic Acid Techniques in Bacterial Systematics John Wiley and Sons Ltd West Sussex England
Stackebrandt E & Goebel BM (1994) Taxonomic note: A place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology Int J Syst Bacteriol 44:846–849
Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH & Swaminathan B (1995) Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: Criteria for bacterial strain typing J Clin Microbiol 33:2233–2239
Williams JKG, Kubelik AR, Livak KJ, Rafalsky JA & Tynger SV (1990) DNA polymorphisms amplified by arbitrary primers are useful as genetic markers Nucleic Acids Res 18:6531–6535
Schwartz DC & Cantor RC (1984) Separation of yeast chromosomeized DNAs by pulse field gradient gel electrophoresis Cell 37:67–75
Regnault B, Grimont F & Grimont PA (1997) Universal ribotyping method using a chemically labelled oligonucleotide probe mixture Res Microbiol 148:649–659
Maslow JN, Mulligan ME & Arbeit RD (1993) Molecular epidemiology: application of contemporary techniques to the typing of microorganism Clin Infect Disease 17:153–164
Olive DM & Bean P (1999) Principles and applications of methods for DNA-based typing of microbial organisms J Clin Microbiol 37:1661–1969
Czekajło KU, Giedrys-Kalemba S & Mędrala D (2006) Phenotypic and genotypic characteristic of Pseudomonas aeruginosa strains isolated from hospitals in the north-west region of Poland. Polish J Microbiol 55:103–112
Versalovic J, Schneider M, de Bruijn FJ, and Lupski JR (1994) Genomic fingerprinting of bacteria using repetitive sequence based PCR (rep-PCR) Meth Cell Mol Biol 5:25–40
Louws FJ, Fulbright DW, Stephens CT & de Bruijn FJ (1995) Differentiation of genomic structure by rep-PCR fingerprinting to rapidly classify Xanthomonas campestris pv. vesicatoria Phytopath 85:528–836
Lupski JR & Weinstock GM (1992) Short, interspersed repetitive DNA sequences in prokaryotic genomes J Bacteriol 174:4525–4529
Stern MJ, Ames GFL, Smith NH, Robinson EC, Higgins CF (1984) Repetitive extragenic palindromic sequences: a major component of the bacterial genome Cell 37:1015–1026
Hulton CSJ, Higgins CF, Sharp PM (1991) ERIC sequences: a novel family of repetitive elements in the genomes of Escherichia coli, Salmonella typhimurium and other enterobacteria Mol Microbiol 5:825–762
Martin B, Humbert O, Camara M, Guenzi E, Walker J, Mitchell T, Andrew P, Prudhomme M, Alloing G, Hakenbeck R, Morrison DA, Boulnois GJ, Claverys J-P (1992) A highly conserved repeated DNA element located in the chromosome of Streptococcus pneumoniae Nucl Acids Res 20:479–3483
Versalovic J, Koeuth T & Lupski, JR (1991) Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucl Acids Res 19:6823–6831
Toth IK, Avrova AO & Hyman LJ (2001) Rapid identification and differentiation of the soft rot Erwinias by 16S–23S Intergenic transcribed spacer-PCR and restriction fragment length polymorphism analyses Appl Environ Microbiol 67:4070–4076
Woese CR, Stackebrandt E, Macke TJ & Fox GE (1985) A phylogenetic definition of major eubacterial taxa Syst Appl Microbiol 6:143–151
Dubnau D, Smith I, Morell P & Marmur J (1965) Gene conservation in Bacillus species I. Conserved genetic and nucleic acid base sequence homologies Proc Natl Acad Sci USA 54:491–498
Pal R, Bala S, Dadhwal M, Kumar M, Dhingra G, Prakash O, Prabagaran SR, Shivaji S, Cullum J Holliger C & Lal R (2005) Hexachlorocyclohexane-degrading bacterial strains Sphingomonas paucimobilis B90A, UT26 and Sp+, having similar lin genes, represent three distinct species, Sphingobium indicum sp. nov., Sphingobium japonicum sp. nov. and Sphingobium francense sp. nov., and reclassification of [Sphingomonas] chungbukensis as Sphingobium chungbukense comb. nov. Int J Syst Evol Microbiol 55:1965–1972
Yang Z (1996) Phylogenetic analysis using parsimony and likelihood methods J Mol Evol 42:294–307
Ludwig W & Schleifer K H (1999) Phylogeny of bacteria beyond the 16S rRNA standard ASM News 65:752–757
Ludwig W, Strunk O, Klugbauer S, Klugbauer N, Weizenegger M, Neumaier J, Bachleitner M & Schleifer KH (1998) Bacterial phylogeny based on comparative sequence analysis Electrophoresis 19:554–568
Stackebrandt E & Liesack W (1993) Nucleic acid and classification In Goodfellow M & O’Donnell AG (Eds.) Handbook of new bacterial systematics. Academic Press Ltd. London pp 151–194
Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky M I, Moore LH, Moore WEC, Murray RGE, Stackebrandt E, Starr MP & Truper HG (1987) International committee on systematics bacteriology. Report of the ad hoc committee on reconciliation of approaches to bacterial systematics Int J Syst Microbiol 37:463–464
Mora RR (2006) DNA-DNA reassociation methods applied to microbial taxonomy and their critical evaluation In Stackebrandt E (Eds), Molecular Identification, Systematics, and Population Structure of Prokaryotes Springer-Verlag Berlin Heidelberg pp. 23–49
Stackebrandt E (2003) The richness of prokaryotic diversity: there must be a species somewhere Food Technol Biotechnol 41:7–22
Broekhuijsen M, Larsson P, Johansson A, Byström M, Eriksson U, Larsson E, Prior RG, Sjöstedt A, Titball RW & Forsman M (2003) Genome-wide DNA microarray analysis of Francisella tularensis strains demonstrates extensive genetic conservation within the species but identifies regions that are unique to the highly virulent F. tularensis subsp. Tularensis. J Clin Microbiol 41:2924–2931
Goodfellow M & O’Donnell AG (1993) Roots of bacterial systematics In Goodfellow, M. & O’Donnell, A. G. (Eds.), Handbook of new bacterial systematics. Academic Press Ltd. London pp. 3–54
Schleifer KH & Kandler O (1972) Peptidoglycan types of bacterial cell walls and their taxonomic implications Bacteriol Rev 36:407–477
Suzuki K, Goodfellow M & O’Donnell AG (1993) Cell envelopes and classification In Goodfellow M & O’Donnell AG (Eds.) Handbook of New Bacterial Systematics Academic Press Ltd London pp. 195–250
Busse J & Auling G (1988) Polyamine pattern as a chemotaxonomic marker within the proteobacteria. Syst Appl Microbiol 11:1–8
Bishop DHL, Pandya KP & King HK (1962) Ubiquinone and vitamin K in bacteria Biochem J 83:606–614
Collins MD & Dorothy J (1981) Distribution of isoprenoid quinone structural types in bacteria and their taxonomic implications Microbiol Reviews 45:316–354
On SLW & Holmes B (1991) Reproducibility of tolerance tests those are useful in the identification of compylobacteria J Clin Microbiol 29:1785–1788
On SLW & Holmes B (1992) Assessment of enzyme detection tests useful in identification of campylobacteria 30:746–749
Colwell RR, Austin B (1981) Numerical taxonomy, In Gerhardt, T. P., Murray, R. G. E., Costilow, R. N., Nester, E. W., Wood, W. A., Krieg, N. R. & Phillips, G. B. (Eds.), Manual of Methods for General Bacteriology. American Society for Microbiology, Washington, DC pp. 444–449
Sneath P (1984) Numerical taxonomy 1 In Krieg, N. R. and Holt, J. G. (Eds.), Bergey’s manual of systematics bacteriology The Williams & Wilkins Co Baltimore pp111–118
Bala S, Khanna R, Dhadwal M, Prabhagaran SR, Shivaji S, Cullum J & Lal R (2004) Reclassification of Amycolatopsis mediterranei 46095 as Amycolatopsis rifamycinica. Int J Syst Evol Microbiol 54:1145–1149
Majumdar S & Lal R (2006) Reclassification of Amycolatopsis orientalis DSM 43387 as Amycolatopsis benzoatilytica sp. nov. Int J Syst Evol Microbiol 56:199–204
Prakash O & Lal R (2006) Description of Sphingobium fuligis sp. nov. a phenanthrene-degrading bacterium from a fly ash site dumping site and reclassification of Sphingomonas cloacae as Sphingobium cloacae comb. nov. Int J Syst Evol Microbiol 56: 2147–2152
Prakash O, Kumari K & Lal R. 2007. Pseudomonas delhiensis sp. nov., from a fly ash dumping site of a thermal power plant Int J Syst Evol Microbiol 57:527–531
Fox GE, Wisotzkey JD & Jurtshuk P Jr (1992) How close is close: 16S rRNA sequence identity may not be sufficient to guarantee species identity Int J Syst Bacteriol 42:166–170
Vandamme P, Harrington CS, Jalava K & On SLW (2000) Missidentifying helicobacters: the Helicobacter cinaedi example J Clin Microbiol 38:2261–2266
Bellemare G, Vigne R, Jordon BR (1973) Interaction between Escherichia coli ribosomal proteins and 5S RNA molecules: recognition of prokaryotic 5S RNAs and rejection of eukaryotic 5S RNAs. Biochimie 55:340–350
Nomura M, Traub P & Bechmann H (1968) Hybrid 30S ribosomal particles reconstituted from components of different bacterial origins Nature 219: 793–799
Wrede P & Erdmann VA (1973) Activities of B. stearothermophilus 50S ribosomes reconstituted with prokaryotic ad eukaryotic 5S RNA. FEBS Lett 33:315–319
Daya-Grosjean L, Geisser M, Stoffler G & Garret RA (1973) Heterologous protein-RNA interactions in bacterial ribosomes. FEBS Lett 37:17–20
Wang Y, Zhang Z, Ramanam N (1997) The actinomycete Thermobispora bispora contains two distinct types of transcriptionally active 16S rRNA genes. J Bacteriol 179:3270–3276
Asai T, Zaporojets D, Squires C & Squires CL (1999) An Escherichia coli strain with all chromosomal rRNA operons inactivated: complete exchange of rRNA genes between bacteria. Proc NatI Acad Sci USA, 96:1971–1976
Yap WH, Zhang Z & Wang Y (1999) Distinct type of rRNA operons exist in the genome of the actinomycete Thermomonospora chromogena and evidence for horizontal transfer of an entire rRNA operon J Bacteriol 181:5201–5209
Mylvaganam S & Dennis PP (1992) Sequence heterogeneity between the two genes encoding 16S rRNA from the halophilic archaebacterium Haloarcula marismortui. Genetics 130:399–410
Dennis PP, Ziesche S & Mylvaganam S (1998) Transcription analysis of two disparate rRNA operons in the halophilic archaeon Haloarcula marismortui J Bacteriol 180:4804–4813
Binnewies TT, Motro Y, Hallin P F, Lund O, Dunn D, La T Hampson D J, Bellgard M, Wassenaar T M & Ussery D W (2006) ten years of bacterial genome sequencing: comparative — genomics — based discoveries Funct Integr Genomics 6:165–185
Perna NT, Plunkett III G, Burland V, Mau B, Glasner JD, Rose DJ, Mayhew GF, Evans PS, Gregor J, Kirkpatrick HA, Posfal G, Hackett J, Klink S, Boutin A, Shao Y, Miller L, Grotbeck EJ, Davis NW, Lim A, Dimalanta ET, Potamousis KD, Apodaca J, Anantharaman TS, Lin J, Yen G, Schwartz DC, Welch RA & Blattner FR (2001) Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529–533
Daubin V, Gouy M, Perrie’re G (2002) A phylogenomic approach to bacterial phylogeny: Evidence of a core of genes sharing a common history. Genome Res 12:1080–1090
Lerat E, Daubin V, Moran NA (2003) From gene trees to organismal phylogeny in prokaryotes: The case of the gamma-proteobacteria. PLoS Biol 1:101–109
Olendzenski L Zhaxybayeva O & Gogarten JP (2002) What’s in a tree? Does horizontal gene transfer determine microbial taxonomy? In: Seckbach J (ed) Symbiosis Dordrecht, The Netherlands Kluwer pp. 63–78
Ochman H, Lawrence JG & Groisman EA (2000) Lateral gene transfer and the nature of bacterial innovation Nature 405:299–304
Kunin V, Goldovsky L, Darzentas N & Ouzounis C A (2005) The net of life: Reconstructing the microbial phylogenetic network Genome Research 15: 954–959
Edward PR, Ewing WH (1986) Identification of Enterobacteriaceae 4th ed. Elsevier Science Publishing Co. Inc., New York
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Prakash, O., Verma, M., Sharma, P. et al. Polyphasic approach of bacterial classification — An overview of recent advances. Indian J Microbiol 47, 98–108 (2007). https://doi.org/10.1007/s12088-007-0022-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12088-007-0022-x