Abstract
Purpose
To study the detailed mechanisms of tumorigenesis and clinical outcomes of centrosomal protein 55 (CEP55) overexpression in renal cell carcinoma.
Materials and methods
Microarray analysis was performed to explore differentially expressed genes in five pairs of RCC tissues. Data of CEP55 expression and corresponding clinical information for 532 RCC patients of TCGA database were downloaded from cBioPortal. The expression of CEP55 in RCC tissues and cells was determined by real-time quantitative reverse transcription PCR (qRT-PCR), Western blot analysis and immunohistochemistry (IHC). Cells were transfected with siRNAs or lentivirus to regulate the expression of CEP55. The effects of CEP55 on proliferation, migration, invasion and epithelial-to-mesenchymal transition (EMT) of RCC cells were determined by MTS, migration and invasion assay and Western blot analysis.
Results
CEP55, one of the most upregulated genes in microarray analysis, was overexpressed in RCC tissues and cells. CEP55 expression was significantly correlated with poor outcome including neoplasm disease stage, histologic grade and TNM status, as well as survival status of patients. In vitro experiments showed that downregulation of CEP55 could dramatically inhibit RCC cell proliferation, migration and invasion, while overexpression of CEP55 could promote these biological behaviors. We further demonstrated that CEP55 knockdown suppressed epithelial–mesenchymal transition (EMT), which was mediated via upregulation of E-cadherin and downregulation of N-cadherin and ZEB1, through PI3K/AKT/mTOR pathway. In contrast, overexpression of CEP55 could promote EMT in RCC cells via the activation of PI3K/AKT/mTOR pathway. Importantly, inhibition of PI3K/AKT/mTOR pathway reduced the effects of CEP55 on the migration, invasion and EMT of RCC cells.
Conclusion
Our study showed that CEP55 could promote EMT through PI3K/AKT/mTOR pathway and might be an effective prognostic marker in RCC.
Similar content being viewed by others
Introduction
Renal cell carcinoma (RCC) is a malignant cancer of the tubular cells of the kidney, which accounts for 2–3% of adult malignant tumors and its morbidity and mortality are still increasing [1]. Although most incidentally detected lesions are small low-grade tumors, up to 17% of all renal cell carcinomas have distant metastases at the time of diagnosis [2]. However, there is no effective treatment for metastatic renal cell carcinoma. Interferon-α (INF-α) and interleukin-2 (IL-2), which were the standard for the treatment of metastatic RCC, fail to provide satisfactory clinical benefit because of their known significant toxicity and modest overall response rates (ORRs) [3]. Molecular-targeted therapy is one of the major strategies for the treatment of metastatic RCC. Several molecular-targeted therapies such as the mTOR inhibitor temsirolimus and the VEGF inhibitor bevacizumab have been proved to improve the clinical outcomes of RCC patients [4, 5]. Because of the complexity of the pathogenesis of malignant tumors, molecular-targeted therapies still have the limitations including low efficiency and drug resistance [6]. In the past decades, although several breakthroughs have been achieved in the study of RCC, the underlying mechanisms of metastasis in renal cell carcinoma remain unclear. Therefore, it is important to explore the mechanism of metastasis and find a new molecular target for RCC.
CEP55 has been initially identified as an important component of abscission, participating in the separation of two daughter cells [7]. Currently, it has been found that CEP55 serves a pivotal role in multiple cancers, such as oral squamous cell carcinoma and breast cancer [8, 9]. Previous studies reported that neoplasms with high levels of CEP55 are typically associated with patients’ clinical characteristics and poor outcomes. CEP55 not only could promote the proliferation of cancer cells, but also promotes invasion and metastasis via the increasing of FOXM1 and the activation of MMP-2 in oral squamous cell carcinoma [9, 10]. However, the detailed mechanisms of tumorigenesis and clinical outcomes of CEP55 overexpression in renal cell carcinoma remain unclear.
Epithelial–mesenchymal transition (EMT) is initially discovered in embryonic development, a shift from epithelial states toward mesenchymal states, accompanying the alteration of adhesion molecules expressed on the cell surfaces [11]. E-cadherin is the most commonly used marker for the epithelial trait and N-cadherin is used for the mesenchymal trait [12]. Many researches reveal that EMT is one of the important cellular phenomena that promotes tumor metastasis. Tumor cells bearing low epithelial phenotype and high mesenchymal phenotype have been referred to more flexible and less stable, with the tendency toward metastasis [13]. Snail, slug and zinc finger E-box binding homeobox 1 (ZEB1) are the key transcription factors that could induce EMT by regulating E-cadherin and N-cadherin expression directly or indirectly [14]. In our study, we found that CEP55 could regulate the process of EMT in RCC. Furthermore, CEP55 promoted cell migration and invasion by activation of PI3KAKT/mTOR pathway. These results suggest a novel CEP55 function in metastasis and might serve as a potential therapeutic target for RCC.
Materials and methods
Patient tissue samples
The RCC tissues and adjacent normal tissues were obtained from patients who underwent surgery only in Sun Yat-Sen Memorial Hospital, Sun Yat-Sen University. Five paired samples were used for mRNA microarray analysis and ten paired samples were used for validation. RCC tissues from 57 patients were used for IHC staining.
IHC staining
IHC staining was performed using a Dako Envision System (Dako, Santa Clara, CA, USA) following the manufacturer’s protocol. Sections were blocked using serum-free protein block buffer (Dako, Santa Clara, CA, USA) for 30 min, after which they were incubated with anti-CEP55 (1:200, Atlas Antibodies, Stockholm, Sweden). The tissue sections were graded as high expression when 10% of tumor cells showed immunopositivity. Biopsies with less than 10% tumor cells showing immunostaining were considered low expression.
Cell culture
The human renal cell carcinoma cell lines ACHN and 786-O were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA), which were cultured in RPMI 1640 culture media supplement (Invitrogen, Carlsbad, CA, USA) with 10% fetal bovine serum (FBS, Sigma, St. Louis, MO, USA). The cancer cell lines were cultured at 37 °C in a humidified atmosphere of 5% CO2.
Microarray analysis
5 paired samples were used for microarray work, which were performed by CapitalBio Technology, China. Hybridized with the Agilent human mRNAs Array V2.0, more than 29,000 mRNA were detected. A fold change of ≥ 2.0 and P ≤ 0.05 was set as the threshold for statistically significant up and downregulated genes, respectively.
RNA interference and cell transfection
siRNAs targeting CEP55 (si#1: 5′-GGGAGAAATTGCACACTTAtt-3′; si#2: 5′-GGACTTTTAGCAAAGATCTtt-3′ [9]) were constructed by Genepharma (Suzhou, China). Lipofectamine RNAiMAX (Thermo Fisher Scientific, MA, USA) was used for transient transfection. All functional assays were conducted 48 h after siRNA transfection. Two types of lentivirus plasmids Lv-CEP55 and Lv-vector (blank vector), conducted from Genechem (Shanghai, China) were transfected into RCC cells to establish the overexpressed group and the control group.
MTS assay
The cell proliferation was measured by MTS assay (CellTiter 96® AQueous one Solution Cell proliferation Assay, Promega, Madison, WI, USA). 1 × 103 cells per well were seeded into 96-well plates. 20 μl MTS solution was added into each well and then incubated at 37 °C, protected from light for 4 h. Optical density was measured at 490 nm. The assay was conducted at 0 h, 24 h, 48 h, 72 h, 96 h and 120 h to generate a growth curve.
Colony formation assay
1 × 103 cells per well were seeded into six-well plates and cultured for 2 weeks to form optimal colonies. Colonies were fixed by paraformaldehyde, stained by crystal violet and then photographed.
Wound healing assay
Cells were seeded in six-well plates and cultured to produce a confluent monolayer. Wound areas were scraped using 200-μl pipette tips. Washed three times with PBS to remove debris and then adding RPMI 1640 culture media without FBS. Wound closure was observed and photographed at 0–24 h under an inverted microscope.
Migration and invasion assay
In migration assay, 1 × 105 cells were suspended in 200 μl serum-free medium for per well and seeded into the upper chamber and RPMI 1640 culture media with 10% FBS was added into the lower chamber. In invasion assay, 1 × 105 cells suspended in 200 μl serum-free medium were seeded into the Matrigel-coated upper chambers. Migratory cells through the polycarbonate membrane after 6 h, and invasive cells after 48 h were fixed by paraformaldehyde, stained by crystal violet and counted at 100 × magnification in at least five randomly selected fields.
Real-time quantitative reverse transcription PCR (qRT-PCR) analysis
Total RNA was isolated from cells by RNAiso Plus (Takara, Kusatsu, Japan). Reverse transcription reaction was carried out with the program for 15 min at 37 °C, and then for 5 s at 85 °C. Quantification of CEP55 and GAPDH was performed with the SYBR® PrimeScript™ RT-PCR Kit (Takara, Kusatsu, Japan). The following primer sets for CEPEE and GAPDH were used: CEP55 primers forward: 5′-TTGGAACAACAGATGCAGGC-3′, reverse: 5′-GAGTGCAGCAGTGGGACTTT-3′; GAPDH primers forward: 5′-GGGAGCCAAAAGGGTCAT-3′, reverse: 5′-GAGTCCTTCCACGATACCAA-3′. Relative quantification was performed with the 2−∆∆Ct method.
Western blot analysis
Total protein was isolated from cells by RIPA lysis buffer and separated by polyacrylamide gels, and then transferred onto PVDF membranes. After blocked with 5% BSA solution, the membranes were incubated with primary antibodies, including CEP55, E-cadherin, N-cadherin, ZEB1 (1:1000, Santa Cruz Biotechnology, CA, USA), PI3K, and mTOR Pathway Antibody Sampler Kit (1:1000, CST, Boston, MA, USA). Membranes were incubated with secondary antibodies for 1 h at room temperature. Protein bands were treated with chemiluminescent HRP Substrate (Millipore Corporation, Billerica, MA, USA) and photographed.
Data downloaded from the cancer genome atlas (TCGA)
Data of CEP55 expression and corresponding clinical information for 532 RCC patients from TCGA database were downloaded from cBioPortal (http://www.cbioportal.org/ [15, 16]), provided visualization, analysis and download of large-scale cancer genomics data sets.
Statistical analysis
Statistical analysis was done by the software SPSS 18.0.0. Quantitative variables are presented as the mean ± SD. The significance of the quantitative PCR results from patient tissues and cell lines, the western blot results measured by ImageJ, and the vitro assays were determined by Student’s t test (two-tailed). The significance of clinical characteristics was determined by t test and χ2 test. The Kaplan–Meier method and log-rank test were used to evaluate the correlation between CEP55 expression and patients’ survival. P < 0.05 was considered statistically significant.
Results
Upregulation of CEP55 in renal cell carcinoma tissues and cells
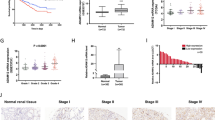
To explore the differentially expressed genes in RCC, a mRNA microarray was utilized to compare mRNA expression profiles between five pairs of renal cell carcinoma tissues and adjacent normal tissues. Hierarchical clustering showed that 267 mRNAs were upregulated in RCC with a fold change > 5, while 200 mRNAs were downregulated with a fold change > 5 (Fig. 1a, P < 0.05). Then, we aimed to identify some functional mRNAs that were upregulated in RCC. Top 10 upregulated mRNAs were chosen for further studies and validated in the paired tissues of ten independent cases. The results showed that CEP55, one of the most upregulated mRNAs in our microarray analysis (fold change 26.2, P < 0.01), was the most overexpressed mRNA in validation (Fig. 1b).
Upregulation of CEP55 in renal cell carcinoma tissues and cells. a Heat map represents unsupervised hierarchical clustering of mRNAs expression level in RCC tissues compared with adjacent normal tissues (ANT). Red color scale represents higher expression level and green color represents lower expression level, b mRNA expression of CEP55 in ten paired RCC tissues. PCR values were normalized to the levels of GAPDH, c mRNA and protein expression of CEP55 in RCC cells. *P < 0.05
We further examined the mRNA and protein expression of CEP55 in RCC cells (Caki-2, 786-O, ACHN and 769-P) and an immortalized normal renal tubular epithelial cell line HK-2. We found that CEP55 was overexpressed in RCC cells compared with HK-2 cells (Fig. 1c), which suggested that the abnormal expression level of CEP55 might be correlated with malignancy.
CEP55 expression is associated with advanced clinicopathological parameters and poor survival
We then analyzed the relationship between CEP55 mRNA expression and clinicopathological parameters of 532 patients from TCGA database. The result was summarized in Tables 1 and 2. Correlation analysis showed that high CEP55 expression level was significantly correlated to higher historical grade (P < 0.01), neoplasm disease stage (P < 0.01) and T stage (P < 0.01). Moreover, we found that expression of CEP55 was positively related to M stage (P < 0.01) and N stage (P < 0.01).
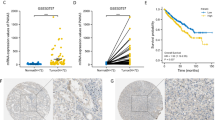
Furthermore, we evaluated the protein expression of CEP55 in 57 pairs of RCC samples. Immunohistochemistry (IHC) staining indicated that CEP55 usually localized in the cytoplasm and slightly in the nucleus of RCC cells. Among 57 RCC samples tested, 35 (61.4%) of them were detected as high expression of CEP55 and 22 (38.6%) were considered as low expression of CEP55 (Fig. 2a). Moreover, immunohistochemistry analysis showed that high expression of CEP55 was associated with advanced neoplasm disease stage and high T stage of RCC (Table 3).
CEP55 expression is associated with advance clinicopathological parameters and poor survival. a Representative images of IHC for CEP55 in clinical RCC samples. Original magnification, × 400, b Kaplan–Meier survival analysis of patients (n = 532) stratified by median CEP55 expression in relation to overall survival. Data were acquired from TCGA database
According to the median mRNA expression of CEP55, the cohort from TCGA was divided into high-expression group (n = 265) and low-expression group (n = 267). There were 175 overall mortality events in this cohort. The significance of the survival risk between low-expression group and high-expression group was assessed with the Kaplan–Meier analysis. As expected, patients in high-expression group had a poorer overall survival compared with patients in low-expression group (Fig. 2b, log-rank test P < 0.01).
CEP55-enhanced cell proliferation and colonization in RCC cells
To study the potential oncogenic role of CEP55 in RCC, loss-of-function studies were performed. To downregulate CEP55 in RCC cells, two small interference RNAs were designed (si#1 and si#2). In both ACHN and 786-O, CEP55 expression levels were effectively reduced with the transfection of si#1 and si#2, respectively (Fig. 3a, b). We next performed a cell proliferation assay and the results revealed that CEP55 knockdown dramatically reduced the proliferative ability of RCC cells compared with the siRNA scramble control (Fig. 3c, P < 0.05). In addition, colony formation assays were performed and the result showed knockdown of CEP55 significantly inhibited colony formation capacity of ACHN and 786O (Fig. 3d).
CEP55-enhanced cell proliferation and colonization in RCC cells. a, b The RNA and protein expression of CEP55 in ACHN and 786-O cells were detected, c CEP55 knockdown inhibited the proliferation of ACHN and 786-O cells, d colony formation with CEP55 knockdown and overexpression in ACHN and 786-O. All cell assays were performed in triplicate. *P < 0.05
To confirm the effect of CEP55 on cell proliferation and colonization in RCC cells, we next transfected CEP55-overexpressed lentivirus into ACHN and 786-O, and stably increased CEP55 expression levels in RCC cell lines (Fig. 3b). Then we found that CEP55 overexpression significantly increased colony formation capacity of ACHN and 786-O (Fig. 3d). However, upregulation of CEP55 had no significant effect on proliferation of ACHN and 786-O. These results suggested that CEP55 promoted cell proliferation and colonization capacity in RCC.
CEP55 promotes migration and invasion in RCC cells
Given that higher CEP55 expression was positively associated with metastasis status in RCC patients, we performed wound healing assay and transwell assay to assess whether CEP55-influenced cell migration and invasion in RCC. Our result demonstrated that the migration and invasion capacity of ACHN and 786-O were significantly weakened after CEP55 silencing, while enhanced after overexpressing (Fig. 4a, b). Taken together, CEP55 promoted migration and invasion of RCC cells.
CEP55 promoted the migration and invasion of RCC cells through EMT
Cell morphology change was observed in overexpressed-CEP55 ACHN cells. Upregulation of CEP55 of ACHN, compared with the empty vector, caused the deformation from a more rounded structure to a more spindle shape (Fig. 5a). It seems that higher CEP55 expression was associated with more elongated and more migratory. Therefore, we attempted to examine the role of CEP55 in EMT of RCC cell lines. The result showed that CEP55 knockdown increased the expression of epithelial marker E-cadherin, and reduced the expression of mesenchymal marker N-cadherin. The protein level of EMT-related transcription factors (EMT-TFs) ZEB1 was decreased significantly after CEP55 knockdown, while snail and slug had no significant change (Fig. 5b). Further research showed that CEP55 overexpression had the motivational effect on EMT with the upregulating N-cadherin and ZEB1 and downregulating E-cadherin (Fig. 5c). Our founding suggested that CEP55 could significantly promote migration and invasion of RCC cells through EMT.
CEP55 promoted the migration and invasion of RCC cells through EMT. a Representative images showing morphological changes in the CEP55-overexpressed ACHN cells compared to the control, b, c western blot analysis for EMT molecular markers in ACHN and 786-O cells with CEP55 knockdown and overexpression
CEP55 induces activation of PI3K/AKT/mTOR pathway in RCC cells
Various evidence showed that the PI3K/AKT/mTOR pathway regulated critical functions including proliferation, migration, invasion, survival and EMT in RCC [17]. Therefore, we evaluated whether CEP55 influenced the PI3K/AKT/mTOR pathway. As a result, we found that knockdown of CEP55 reduced the expression of PI3K and mTOR, as well as the phosphorylation of AKT-Ser473 and mTOR-Ser2448 concurrently (Fig. 6a). In contrast, upregulation of CEP55 induced the activation of PI3K/AKT/mTOR in ACHN and 786-O, compared with vehicle control cells (Fig. 6b).
Inhibition of PI3K/AKT/mTOR pathway reduced the effects of CEP55 on the migration, invasion and EMT of RCC cells
To verify whether CEP55 promoted EMT of RCC cell lines through the activation of PI3K/AKT/mTOR pathway, the PI3K-specific inhibitor LY294002 was added into the CEP55-overexpressed cell to observe whether the pro-EMT effect of CEP55 could be waken. As expected, CEP55-induced AKT-Ser473 phosphorylation were effectively abolished in ACHN and 786-O incubated with LY294002 at the final concentration of 10 μM for 2 h. Then, depressed cell migration and invasion was observed in ACHN and 786-O with incubation of LY294002 (Fig. 7a). Moreover, LY294002 also attenuated the pro-EMT effect driven by CEP55, including the increasing of N-cadherin and ZEB1, as well as the decreasing of E-cadherin (Fig. 7b). These results showed that CEP55 promoted cell migration and invasion by activating EMT through PI3K/AKT/mTOR pathway.
Inhibition of PI3K/AKT/mTOR pathway reduced the effects of CEP55 on the migration, invasion and EMT of RCC cells. a Migration and invasion assay for CEP55-overexpressed RCC cells treated with DMSO or LY294002, b Western blot analysis for phosphorylated form of AKT and mTOR, CEP55 and EMT-related proteins treated with DMSO or LY294002. *P < 0.05
Discussion
Previous studies have shown that CEP55 is aberrantly expressed in multiple types of cancers, and involved in cancer progression, including proliferation, migration and invasion [18]. CEP55 knockdown could suppress cellular proliferation as a result of cell cycle arrest at the G2/M phase in gastric cancer [19]. In oral cavity squamous cell carcinoma, CEP55 could promote MMP-2 expression and activation through upregulation of FOXM1, serving as a driver of invasion and metastasis [9]. In this study, we investigated the expression level of CEP55 in RCC tissues and adjacent normal tissues, and a panel of RCC cell lines. The expression of CEP55 mRNA was significantly associated with poor survival and advanced features of patients with RCC, including neoplasm disease stage, histologic grade and TNM stage. The results suggested that CEP55 might function as an oncogene in RCC. To elucidate whether CEP55 affects the biological behavior of RCC cells, we designed a series of experiments to evaluate the biological function of CEP55. Then we found that knockdown of CEP55 could significantly decrease the proliferation, migration and invasion of RCC cells. In contrast, overexpression of CEP55 enhanced the capacity of migration and invasion in RCC cells.
Emerging evidence has shown that EMT is a critical component of the metastatic process, leading the dissociation of connected cells from the primary tumor and their intravasation into blood vessels [20, 21]. Loss of epithelial marker expression and gain of the mesenchymal marker expression are the hallmarks of the EMT process. EMT-TF such as snail, slug and ZEB1 are the spotlight of EMT investigation [13]. Upregulation of ZEB1 has been observed in various cancers and the expression of ZEB1 is significantly correlated to tumor grade and lymphatic or distant metastasis [22,23,24]. Previous studies showed that ZEB1 induces EMT by binding to the promoter of E-cadherin and depressing the expression level of E-cadherin. In addition, it is known that miRNA-200 acts as a regulator of E-cadherin and plays a crucial role in modulating EMT in RCC. ZEB1 has also been proven to inhibit E-cadherin by forming an important negative feedback loop with miRNA-200 [25, 26]. In the present study, our data indicated that knockdown of CEP55 could increase the expression of E-cadherin and decrease the expression of N-cadherin and ZEB1. In addition, after upregulation of CEP55 expression, the expression of N-cadherin and ZEB1 was increased and E-cadherin was decreased in RCC cells. These findings suggest that CEP55 may be an EMT inducer and has potential as a novel molecular target for anti-metastasis therapy in RCC.
Increasing evidence shows that the activation of PI3K/AKT/mTOR pathway is crucial for EMT process [27, 28]. Mechanistically, activated PI3K could phosphorylate phosphatidylinositol 4, 5-bisphosphate (PIP2), which would bind and phosphorylate AKT [29]. Furthermore, phosphorylation of AKT could activate mTOR to control motility, invasion and phenotype transformation of cells and contribute to EMT [30]. Previous study showed that miR-200c suppresses the expression of ZEB1 through PI3K/AKT signaling pathway in non-small cell lung cancer [31]. Furthermore, Src promotes the process of EMT by upregulation of ZEB1 and ZEB2 through AKT signaling pathway in gastric cancer cells [32]. In addition, it has been reported that CEP55 binds to PI3K and increases the stability of this subunit, resulting in increased AKT activation as observed by an increase in S473 phosphorylation [33]. In our study, we found that activation of PI3K/AKT/mTOR pathway was positively associated with the expression of N-cadherin and ZEB1 and negatively associated with the expression of E-cadherin. In addition, after inhibiting PI3K with LY294002, the expression of E-cadherin was upregulated while N-cadherin and ZEB1 was downregulated. Therefore, it came to the conclusion that CEP55 activated the PI3K/AKT/mTOR pathway and participated in EMT process of RCC.
At present, molecular target therapy is the first option for the management of advanced RCC. Multitarget tyrosine kinase inhibitors (TKIs) which against the VEGF receptors (VEGFRs), PDGF receptors (PDGFRs) have already been shown to improve survival of RCC patients [34]. A randomized, placebo-controlled, phase 3 study showed that mTOR inhibitor everolimus could improve the median progression-free survival (PFS) of advanced RCC patients [35]. However, the toxicity and resistance has limited their clinical application. Activation of alternative or compensatory pathway of VEGFR and mTOR signaling contributes to the upregulation of HIF, which leads to the resistance to VEGFR- and mTOR-targeted therapies [36]. Due to the negative feedback loop between mTOR, PI3K and MAPK, inhibition of mTOR would lead to the activation of PI3K/AKT and MAPK pathway [37]. In our study, CEP55 inhibition could simultaneously weaken PI3K/AKT/mTOR pathway, so CEP55 may be an effective target for metastasis RCC treatment.
Abbreviations
- CEP55:
-
Centrosomal protein 55
- RCC:
-
Renal cell carcinoma
- EMT:
-
Epithelial–mesenchymal transition
- INF-α:
-
Interferon-α
- IL-2:
-
Interleukin-2
- ORRs:
-
Overall response rates
- ZEB1:
-
Zinc-finger E-box-binding 1
- ANT:
-
Adjacent normal tissue
- IHC:
-
Immunohistochemistry
- EMT-TFs:
-
EMT-associated transcription factors
- OD:
-
Optical density
- qRT-PCR:
-
Real-time quantitative reverse transcription PCR
- FBS:
-
Fetal bovine serum
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30.
Capitanio U, Montorsi F. Renal cancer. Lancet. 2016;387(10021):894–906.
Barata PC, Rini BI. Treatment of renal cell carcinoma: current status and future directions. CA Cancer J Clin. 2017;67(6):507–24. https://doi.org/10.3322/caac.21411.
Hudes G, Carducci M, Tomczak P, Dutcher J, Figlin R, Kapoor A, et al. Temsirolimus, interferon alfa, or both for advanced renal-cell carcinoma. N Engl J Med. 2007;356(22):2271–81. https://doi.org/10.1056/NEJMoa066838.
Escudier B, Pluzanska A, Koralewski P, Ravaud A, Bracarda S, Szczylik C, et al. Bevacizumab plus interferon alfa-2a for treatment of metastatic renal cell carcinoma: a randomised, double-blind phase III trial. Lancet. 2007;370(9605):2103–11. https://doi.org/10.1016/S0140-6736(07)61904-7.
Garcia JA, Rini BI. Recent progress in the management of advanced renal cell carcinoma. CA Cancer J Clin. 2007;57(2):112–25.
Fabbro M, Zhou BB, Takahashi M, Sarcevic B, Lal P, Graham ME, et al. Cdk1/Erk2- and Plk1-dependent phosphorylation of a centrosome protein, CEP55, is required for its recruitment to midbody and cytokinesis. Dev Cell. 2005;9(4):477–88.
Wang Y, Jin T, Dai X, Xu J. Lentivirus-mediated knockdown of CEP55 suppresses cell proliferation of breast cancer cells. Biosci Trends. 2016;10(1):67–73.
Chen CH, Chien CY, Huang CC, Hwang CF, Chuang HC, Fang FM, et al. Expression of FLJ10540 is correlated with aggressiveness of oral cavity squamous cell carcinoma by stimulating cell migration and invasion through increased FOXM1 and MMP-2 activity. Oncogene. 2009;28(30):2723–37.
Chen CH, Shiu LY, Su LJ, Huang CY, Huang SC, Huang CC, et al. FLJ10540 is associated with tumor progression in nasopharyngeal carcinomas and contributes to nasopharyngeal cell proliferation, and metastasis via osteopontin/CD44 pathway. J Transl Med. 2012;10:93.
Hay ED. An overview of epithelio-mesenchymal transformation. Acta Anat (Basel). 1995;154(1):8–20.
Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial–mesenchymal transitions in development and disease. Cell. 2009;139(5):871–90.
Nieto MA, Huang RY, Jackson RA, Thiery JP. EMT: 2016. Cell. 2016;166(1):21–45.
Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial–mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15(3):178–96.
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–4.
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1.
Xu W, Yang Z, Lu N. A new role for the PI3K/Akt signaling pathway in the epithelial–mesenchymal transition. Cell Adhes Migr. 2015;9(4):317–24. https://doi.org/10.1080/19336918.2015.1016686.
Jeffery J, Sinha D, Srihari S, Kalimutho M, Khanna KK. Beyond cytokinesis: the emerging roles of CEP55 in tumorigenesis. Oncogene. 2016;35(6):683–90.
Tao J, Zhi X, Tian Y, Li Z, Zhu Y, Wang W, et al. CEP55 contributes to human gastric carcinoma by regulating cell proliferation. Tumour Biol. 2014;35(5):4389–99.
Ye X, Brabletz T, Kang Y, Longmore GD, Nieto MA, Stanger BZ, et al. Upholding a role for EMT in breast cancer metastasis. Nature. 2017;547(7661):E1–3.
Aiello NM, Brabletz T, Kang Y, Nieto MA, Weinberg RA, Stanger BZ. Upholding a role for EMT in pancreatic cancer metastasis. Nature. 2017;547(7661):E7–8.
Rhim AD, Mirek ET, Aiello NM, Maitra A, Bailey JM, McAllister F, et al. EMT and dissemination precede pancreatic tumor formation. Cell. 2012;148(1–2):349–61.
Takeyama Y, Sato M, Horio M, Hase T, Yoshida K, Yokoyama T, et al. Knockdown of ZEB1, a master epithelial-to-mesenchymal transition (EMT) gene, suppresses anchorage-independent cell growth of lung cancer cells. Cancer Lett. 2010;296(2):216–24.
Han KS, Li N, Raven PA, Fazli L, Ettinger S, Hong SJ, et al. Targeting integrin-linked kinase suppresses invasion and metastasis through downregulation of epithelial-to-mesenchymal transition in renal cell carcinoma. Mol Cancer Ther. 2015;14(4):1024–34.
Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, Wang J, Shannon MF, et al. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial–mesenchymal transition. Cancer Res. 2008;68(19):7846–54.
Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, et al. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep. 2008;9(6):582–9.
Courtney KD, Corcoran RB, Engelman JA. The PI3K pathway as drug target in human cancer. J Clin Oncol. 2010;28(6):1075–83.
Lamouille S, Derynck R. Cell size and invasion in TGF-beta-induced epithelial to mesenchymal transition is regulated by activation of the mTOR pathway. J Cell Biol. 2007;178(3):437–51.
Lamouille S, Derynck R. Emergence of the phosphoinositide 3-kinase-Akt-mammalian target of rapamycin axis in transforming growth factor-beta-induced epithelial–mesenchymal transition. Cells Tissues Organs. 2011;193(1–2):8–22. https://doi.org/10.1159/000320172.
Gonzalez DM, Medici D. Signaling mechanisms of the epithelial–mesenchymal transition. Sci Signal. 2014;7(344):re8.
Zhou G, Zhang F, Guo Y, Huang J, Xie Y, Yue S, et al. miR-200c enhances sensitivity of drug-resistant non-small cell lung cancer to gefitinib by suppression of PI3K/Akt signaling pathway and inhibits cell migration via targeting ZEB1. Biomed Pharmacother. 2017;85:113–9. https://doi.org/10.1016/j.biopha.2016.11.100.
Zhao L, Li X, Song N, Li A, Hou K, Qu X, et al. Src promotes EGF-induced epithelial-to-mesenchymal transition and migration in gastric cancer cells by upregulating ZEB1 and ZEB2 through AKT. Cell Biol Int. 2017;1:1. https://doi.org/10.1002/cbin.10894.
Chen CH, Lu PJ, Chen YC, Fu SL, Wu KJ, Tsou AP, et al. FLJ10540-elicited cell transformation is through the activation of PI3-kinase/AKT pathway. Oncogene. 2007;26(29):4272–83.
Motzer RJ, Hutson TE, Tomczak P, Michaelson MD, Bukowski RM, Rixe O, et al. Sunitinib versus interferon alfa in metastatic renal-cell carcinoma. N Engl J Med. 2007;356(2):115–24. https://doi.org/10.1056/NEJMoa065044.
Motzer RJ, Escudier B, Oudard S, Hutson TE, Porta C, Bracarda S, et al. Phase 3 trial of everolimus for metastatic renal cell carcinoma: final results and analysis of prognostic factors. Cancer. 2010;116(18):4256–65. https://doi.org/10.1002/cncr.25219.
Rini BI, Atkins MB. Resistance to targeted therapy in renal-cell carcinoma. Lancet Oncol. 2009;10(10):992–1000. https://doi.org/10.1016/S1470-2045(09)70240-2.
Carracedo A, Ma L, Teruya-Feldstein J, Rojo F, Salmena L, Alimonti A, et al. Inhibition of mTORC1 leads to MAPK pathway activation through a PI3K-dependent feedback loop in human cancer. J Clin Investig. 2008;118(9):3065–74. https://doi.org/10.1172/JCI34739.
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (Grant nos. 81472388, 81672534), supported by grant [2013]163 from Key Laboratory of Malignant Tumor Molecular Mechanism and Translational Medicine of Guangzhou Bureau of Science and Information Technology, supported by Grant KLB09001 from the Key Laboratory of Malignant Tumor Gene Regulation and Target Therapy of Guangdong Higher Education Institutes, supported by grant from Guangdong Science and Technology Department (2013B021800110, 2015B050501004), supported by Guangdong Natural Science Foundation (2014A030310133).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Informed consent
Informed consent was obtained from each patient on the day of admission.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Haicheng Chen, Dingjun Zhu and Zaosong Zheng contributed equally to this work.
Rights and permissions
OpenAccess This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Chen, H., Zhu, D., Zheng, Z. et al. CEP55 promotes epithelial–mesenchymal transition in renal cell carcinoma through PI3K/AKT/mTOR pathway. Clin Transl Oncol 21, 939–949 (2019). https://doi.org/10.1007/s12094-018-02012-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12094-018-02012-8