Abstract
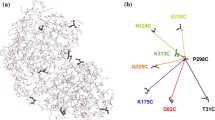
The Hsp60-type chaperonin GroEL assists in the folding of the enzyme human carbonic anhydrase II (HCA II) and protects it from aggregation. This study was aimed to monitor conformational rearrangement of the substrate protein during the initial GroEL capture (in the absence of ATP) of the thermally unfolded HCA II molten-globule. Single- and double-cysteine mutants were specifically spin-labeled at a topological breakpoint in the β-sheet rich core of HCA II, where the dominating antiparallel β-sheet is broken and β-strands 6 and 7 are parallel. Electron paramagnetic resonance (EPR) was used to monitor the GroEL-induced structural changes in this region of HCA II during thermal denaturation. Both qualitative analysis of the EPR spectra and refined inter-residue distance calculations based on magnetic dipolar interaction show that the spin-labeled positions F147C and K213C are in proximity in the native state of HCA II at 20 °C (as close as ∼8 Å), and that this local structure is virtually intact in the thermally induced molten-globule state that binds to GroEL. In the absence of GroEL, the molten globule of HCA II irreversibly aggregates. In contrast, a substantial increase in spin–spin distance (up to >20 Å) was observed within minutes, upon interaction with GroEL (at 50 and 60 °C), which demonstrates a GroEL-induced conformational change in HCA II. The GroEL binding-induced disentanglement of the substrate protein core at the topological break-point is likely a key event for rearrangement of this potent aggregation initiation site, and hence, this conformational change averts HCA II misfolding.





Similar content being viewed by others
Abbreviations
- A ptp :
-
peak to peak amplitude
- EPR:
-
Electron paramagnetic resonance
- FRET:
-
Fluorescence resonance energy transfer
- GuHCl:
-
Guanidine hydrochloride
- HCA II:
-
Human carbonic anhydrase II
- MTSSL:
-
(1-oxyl-2,2,5,5-tetramethylpyrroline-3-methyl)-methanethiosulfonate spin label
- R1:
-
Cysteine side chain labeled with MTSSL
References
Chiti F, Dobson CM (2006) Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem 75:333–366
Lambert MP, Barlow AK, Chromy BA, Edwards C, Freed R, Liosatos M, Morgan TE, Rozovsky I, Trommer B, Viola KL, Wals P, Zhang C, Finch CE, Krafft GA, Klein WL (1998) Diffusible, nonfibrillar ligands derived from Abeta1-42 are potent central nervous system neurotoxins. Proc Natl Acad Sci USA 95:6448–6453
Hammarström P, Persson M, Freskgård PO, Mårtensson LG, Andersson D, Jonsson BH, Carlsson U (1999) Structural mapping of an aggregation nucleation site in a molten globule intermediate. J Biol Chem 274:32897–32903
Wiseman RL, Powers ET, Buxbaum JN, Kelly JW, Balch WE (2007) An adaptable standard for protein export from the endoplasmic reticulum. Cell 131(4):809–821
London J, Skrzynia C, Goldberg ME (1974) Renaturation of Escherichia coli tryptophanase after exposure to 8 M urea. Evidence for the existence of nucleation centers. Eur J Biochem 47:409–415
Campioni S, Mossuto MF, Torrassa S, Calloni G, de Laureto PP, Relini A, Fontana A, Chiti F (2008) Conformational properties of the aggregation precursor state of HypF-N. J Mol Biol 379(3):554–567
Saibil HR, Zheng D, Roseman AM, Hunter AS, Watson GMF, Chen S, auf der Mauer A, O'Hara BP, Wood SP, Mann NH (1993) ATP induces large quaternary rearrangements in a cage-like chaperonin structure. Curr Biol 3:265–273
Zahn R, Perrett S, Fersht AR (1996) Conformational states bound by the molecular chaperones GroEL and secB: a hidden unfolding (annealing) activity. J Mol Biol 261:43–61
Horwich AL, Fenton WA (2009) Chaperonin-mediated protein folding: using a central cavity to kinetically assist polypeptide chain folding. Q Rev Biophys 42(2):83–116
Persson M, Lindgren M, Hammarström P, Svensson M, Jonsson BH, Carlsson U (1999) EPR mapping of interactions between spin-labeled variants of human carbonic anhydrase II and GroEL: evidence for increased flexibility of the hydrophobic core by the interaction. Biochemistry 38:432–441
Hammarström P, Persson M, Owenius R, Lindgren M, Carlsson U (2000) Protein substrate binding induces conformational changes in the chaperonin GroEL. A suggested mechanism for unfoldase activity. J Biol Chem 275:22832–22838
Hammarström P, Persson M, Carlsson U (2001) Protein compactness measured by fluorescence resonance energy transfer. Human carbonic anhydrase II is considerably expanded by the interaction of GroEL. J Biol Chem 276:21765–21775
Villebeck L, Persson M, Luan SL, Hammarström P, Lindgren M, Jonsson BH (2007) Conformational rearrangements of tail-less complex polypeptide 1 (TCP-1) ring complex (TRiC)-bound actin. Biochemistry 46:5083–5093
Villebeck L, Moparthi SB, Lindgren M, Hammarström P, Jonsson BH (2007) Domain-specific chaperone-induced expansion is required for beta-actin folding: a comparison of beta-actin conformations upon interactions with GroEL and tail-less complex polypeptide 1 ring complex (TRiC). Biochemistry 46:12639–12647
Slepenkov SV, Witt SN (2002) The unfolding story of the Escherichia coli Hsp70 DnaK: is DnaK a holdase or an unfoldase? Mol Microbiol 45:1197–1206
Zolkiewski M (2006) A camel passes through the eye of a needle: protein unfolding activity of Clp ATPases. Mol Microbiol 61:1094–1100
Viitanen PV, Donaldson GK, Lorimer GH, Lubben TH, Gatenby AA (1991) Complex interactions between the chaperonin 60 molecular chaperone and dihydrofolate reductase. Biochemistry 30:9716–9723
Zahn R, Perrett S, Stenberg G, Fersht AR (1996) Catalysis of amide proton exchange by the molecular chaperones GroEL and SecB. Science 271:642–645
Reid BG, Flynn GC (1996) GroEL binds to and unfolds rhodanese posttranslationally. J Biol Chem 271:7212–7217
Carlsson U, Jonsson BH (1995) Folding of beta-sheet proteins. Curr Opin Struct Biol 5:482–487
Persson M, Carlsson U, Bergenhem NCH (1996) GroEL reversibly binds to, and causes rapid inactivation of, human carbonic anhydrase II at high temperatures. Biochim Biophys Acta 1298:191–198
Goldberg MS, Zhang J, Sondek S, Matthews CR, Fox RO, Horwich AL (1997) Native-like structure of a protein-folding intermediate bound to the chaperonin GroEL. Proc Natl Acad Sci USA 94:1080–1085
Chen J, Walter S, Horwich AL, Smith DL (2001) Folding of malate dehydrogenase inside the GroEL-GroES cavity. Nat Struct Biol 8:721–728
Shortle D, Ackerman MS (2001) Persistence of native-like topology in a denatured protein in 8 M urea. Science 293:487–489
Gervasoni P, Staudenmann W, James P, Plückthun A (1998) Identification of the binding surface on beta-lactamase for GroEL by limited proteolysis and MALDI-mass spectrometry. Biochemistry 37:11660–11669
Shtilerman M, Lorimer GH, Englander SW (1999) Chaperonin function: folding by forced unfolding. Science 284:822–825
Elad N, Farr GW, Clare DK, Orlova EV, Horwich AL, Saibil HR (2007) Topologies of a substrate protein bound to the chaperonin GroEL. Mol Cell 26:415–426
Hammarström P, Jonsson BH (2005) Protein denaturation and the denatured state. Encyclopedia of Life Sciences. Wiley, New York (doi:10.1038/npg.els.0003003)
Berliner LJ, Eaton SS, Eaton GR (eds) (2001) Biological magnetic resonance, Vol. 19: distance measurements in biological systems by EPR. Kluwer Academic/Plenum, New York
Persson M, Harbridge JR, Hammarström P, Mitri R, Mårtensson LG, Carlsson U, Eaton GR, Eaton SS (2001) Comparison of electron paramagnetic resonance methods to determine distances between spin labels on human carbonic anhydrase II. Biophys J 80:2886–2897
Jeschke G, Polyhach Y (2007) Distance measurements on spin-labelled biomacromolecules by pulsed electron paramagnetic resonance. Phys Chem Chem Phys 9:1895–1910
Fleissner MR, Brustad EM, Kálai T, Altenbach C, Cascio D, Peters FB, Hideg K, Schultz PG, Hubbell WL (2009) Site-directed spin labeling of a genetically encoded unnatural amino acid. Proc Natl Acad Sci USA 106(51):21637–21642
Mårtensson LG, Jonsson BH, Freskgård PO, Kihlgren A, Svensson M, Carlsson U (1993) Characterization of folding intermediates of human carbonic anhydrase II: probing substructure by chemical labeling of SH groups introduced by site-directed mutagenesis. Biochemistry 32:224–231
Hammarström P, Owenius R, Mårtensson LG, Carlsson U, Lindgren M (2001) High-resolution probing of local conformational changes in proteins by the use of multiple labeling: unfolding and self-assembly of human carbonic anhydrase II monitored by spin, fluorescent, and chemical reactivity probes. Biophys J 80:2867–2885
Svensson M, Freskgard PO, Lindgren M, Boren K, Carlsson U (1995) Mapping the folding intermediate of human carbonic anhydrase II. Probing substructure by chemical reactivity and spin and fluorescence labeling of engineered cysteine residues. Biochemistry 34:8606–8620
Altenbach C, Oh KJ, Trabanino RJ, Hideg K, Hubbell WL (2001) Estimation of inter-residue distances in spin labeled proteins at physiological temperatures: experimental strategies and practical limitations. Biochemistry 40:15471–15482
Hammarström P, Carlsson U (2000) Is the unfolded state the Rosetta Stone of the protein folding problem? Biochem Biophys Res Commun 276:393–398
Eriksson EA, Jones AT, Liljas A (1988) Refined structure of human carbonic anhydrase II at 2.0 A resolution. Proteins Struct Funct Genet 4:274–282
Håkansson K, Carlsson M, Svensson LA, Liljas A (1992) Structure of native and apo carbonic anhydrase II and structure of some of its anion-ligand complexes. J Mol Biol 227:1192–1204
Anthony-Cahill SJ, Benfield PA, Fairman R, Wasserman ZR, Brenner SL, Stafford WF, Altenbach C, Hubbell WL, De Grado WF (1992) Molecular characterization of helix-loop-helix peptides. Science 255:979–983
Farrens DL, Altenbach C, Yang K, Hubbell WL, Khorana HG (1996) Requirement of rigid-body motion of transmembrane helices for light activation of rhodopsin. Science 274:768–770
Rabenstein MD, Shin YK (1995) Determination of the distance between two spin labels attached to a macromolecule. Proc Natl Acad Sci USA 92:8239–8243
Altenbach C, Cai K, Klein-Seetharaman J, Khorana HG, Hubbell WL (2001) Structure and function in rhodopsin: mapping light-dependent changes in distance between residue 65 in helix TM1 and residues in the sequence 306-319 at the cytoplasmic end of helix TM7 and in helix H8. Biochemistry 40:15483–15492
Serag AA, Altenbach C, Gingery M, Hubbell WL, Yeates TO (2002) Arrangement of subunits and ordering of beta-strands in an amyloid sheet. Nat Struct Biol 9:734–739
Zhang JH, Xiao G, Gunsalus RP, Hubbell WL (2003) Phosphorylation triggers domain separation in the DNA binding response regulator NarL. Biochemistry 42:2552–2559
Sot B, Bañuelos S, Valpuesta JM, Muga A (2003) GroEL stability and function. Contribution of the ionic interactions at the inter-ring contact sites. J Biol Chem 278(34):32083–32090
Back JF, Oakenfull D, Smith MB (1979) Increased thermal stability of proteins in the presence of sugars and polyols. Biochemistry 18:5191–5196
Lee JC, Timasheff SN (1981) The stabilization of proteins by sucrose. J Biol Chem 256:7193–7201
Mchaourab HS, Oh KJ, Fang CJ, Hubbell WL (1997) Conformation of T4 lysozyme in solution. Hinge-bending motion and the substrate-induced conformational transition studied by site-directed spin labeling. Biochemistry 36:307–316
Mchaourab HS, Lietzow MA, Hideg K, Hubbell WL (1996) Motion of spin-labeled side chains in T4 lysozyme. Correlation with protein structure and dynamics. Biochemistry 35:7692–7704
Owenius R, Österlund M, Lindgren M, Svensson M, Olsen OH, Persson E, Freskgård PO, Carlsson U (1999) Properties of spin and fluorescent labels at a receptor-ligand interface. Biophys J 77:2237–2250
Owenius R, Österlund M, Svensson M, Lindgren M, Persson E, Freskgård PO, Carlsson U (2001) Spin and fluorescent probing of the binding interface between tissue factor and factor VIIa at multiple sites. Biophys J 81:2357–2369
Langen R, Oh KJ, Cascio D, Hubbell WL (2000) Crystal structures of spin labeled T4 lysozyme mutants: implications for the interpretation of EPR spectra in terms of structure. Biochemistry 39:8396–8405
Mathlouthi M, Reiser P (eds) (1995) Sucrose: properties and applications. Blackie, London
Weber A, Schiemann O, Bode B, Prisner TF (2002) PELDOR at S- and X-band frequencies and the separation of exchange coupling from dipolar coupling. J Magn Reson 157:277–285
Fiori WR, Millhauser GL (1995) Exploring the peptide 3(10)-helix<–>alpha-helix equilibrium with double label electron spin resonance. Biopolymers 37:421–431
Sale K, Song L, Liu Y-S, Perozo E, Fajer P (2005) Explicit treatment of spin labels in modeling of distance constraints from dipolar EPR and DEER. J Am Chem Soc 127:9334–9335
Banham JE, Baker CM, Ceola S, Day IJ, Grant GH, Groenen EJJ, Rogers CT, Jeschke G, Timmel CR (2008) Distance measurements in the borderline region of applicability of CW EPR and DEER: a model study on a homologous series of spin-labelled peptides. J Magn Reson 191:202–218
Lührs T, Ritter C, Adrian M, Riek-Loher D, Bohrmann B, Döbeli H, Schubert D, Riek R (2005) 3D structure of Alzheimer's amyloid-beta(1-42) fibrils. Proc Natl Acad Sci USA 102:17342–17347
Ritter C, Maddelein ML, Siemer AB, Lührs T, Ernst M, Meier BH, Saupe SJ, Riek R (2005) Correlation of structural elements and infectivity of the HET-s prion. Nature 435:844–848
Farr GW, Furtak K, Rowland MB, Ranson NA, Saibil HR, Kirchhausen T, Horwich AL (2000) Multivalent binding of nonnative substrate proteins by the chaperonin GroEL. Cell 100:561–573
Llorca O, McCormack EA, Hynes G, Grantham J, Cordell J, Carrascosa JL, Willison KR, Fernandez JJ, Valpuesta JM (1999) Eukaryotic type II chaperonin CCT interacts with actin through specific subunits. Nature 402:693–696
Albert A, Yunta C, Arranz R, Peña A, Salido E, Valpuesta JM, Martín-Benito J (2010) Structure of GroEL in complex with an early folding intermediate of alanine glyoxylate aminotransferase. J Biol Chem 285(9):6371–6376
Acknowledgements
We thank Professor Anders Lund for giving us access to the EPR lab at IFM-Chemical Physics, Linköping University and Professor Sandra S. Eaton, University of Denver, for advice concerning EPR data analysis. We acknowledge Professor Mikael Lindgren and Dr. Malin Persson for valuable discussions and advice. We also gratefully acknowledge Dr. Christian Altenbach, University of California, Los Angeles, for providing his software for determining inter-residue distances. This work was supported by grants from the Swedish Foundation for Strategic Research (PH), the Swedish Research Council (BHJ, PH, and UC), and the Knut and Alice Wallenberg Foundation (BHJ, PH, and UC). PH is a Royal Swedish Academy of Sciences Research Fellow supported by a grant from the Knut and Alice Wallenberg Foundation.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Owenius, R., Jarl, A., Jonsson, BH. et al. GroEL-induced topological dislocation of a substrate protein β-sheet core: a solution EPR spin–spin distance study. J Chem Biol 3, 127–139 (2010). https://doi.org/10.1007/s12154-010-0038-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12154-010-0038-2