Abstract
Background
About 2-7% of familial cardiomyopathy cases are caused by a mutation in the gene encoding cardiac troponin I (TNNI3). The related clinical phenotype is usually severe with early onset. Here we report on all currently known mutations in the Dutch population and compared these with those described in literature.
Methods
TheTNNI3 gene was screened for mutations in all coding exons and flanking intronic sequences in a large cohort of cardiomyopathy patients. All Dutch index cases carrying a TNNI3 mutation that are described in this study underwent extensive cardiological evaluation and were listed by their postal codes.
Results
In 30 families, 14 different mutations were identified. Three TNNI3 mutations were found relatively frequently in both familial and non-familial cases of hypertrophic cardiomyopathy (HCM) or restrictive cardiomyopathy (RCM). Haplotype analysis showed that p.Arg145Trp and p.Ser166Phe are founder mutations in the Netherlands, while p.Glu209Ala is not. The majority of Dutch TNNI3 mutations were associated with a HCM phenotype. Mean age at diagnosis was 36.5 years. Mutations causing RCM occurred less frequently, but were identified in very young children with a poor prognosis.
Conclusion
In line with previously published data, we found TNNI3 mutations to be rare and associated with early onset and severe clinical presentation.
Similar content being viewed by others
Introduction
Inherited hypertrophic cardiomyopathy (HCM) is caused by genetic mutations that affect the cardiac sarcomere in around 75–80% of cases and is transmitted in an autosomal dominant way. Early disease onset (< 45 years of age), familial occurrence and an asymmetrical left ventricular wall thickness of more than 20 mm are globally accepted criteria for the diagnosis of HCM. In patients fulfilling these criteria there is a high likelihood of finding a causative mutation. About 30–65% of these mutations are found in only nine sarcomeric genes which are investigated in most Dutch cardiogenetic centres. The performance of the sarcomere highly depends on a functioning sliding mechanism, explaining why the majority of mutations are found in genes, such as the beta-myosin heavy chain (MYH7) and myosin binding protein-C3 (MYBPC3) gene, encoding proteins which are crucial for this function [1].
Cardiomyocyte contraction is dependent on local sarcomeric calcium levels, which is regulated by the small troponin complex, consisting of the three subunits troponin I, C and T [2]. Troponin I (TNNI3) binds to the thin filament of the sarcomere and prevents muscle contraction by inhibiting the actomyosin activity of the myosin heavy chain. This inhibitory effect is released by calcium binding to troponin C. This triggers a series of events including an altered interaction within the actin–troponin–tropomyosin binding complex, displacement of the myosin head along the thin filament and adenosine triphosphate (ATP) hydrolysis, resulting in force generation. Troponin I3 is exclusively expressed in cardiac muscle and plays a central role in modulating cardiac muscle contraction and relaxation in response to changes in intracellular calcium.
The prevalence of mutations in TNNI3 is believed to be <5% in different types of cardiomyopathies such as hypertrophic (HCM), restrictive (RCM) and dilated (DCM) cardiomyopathy [3]. Phenotypic manifestations range from clinically mild to severe with poor prognosis often caused by sudden cardiac death [3, 4]. In the literature, the majority of mutations in TNNI3 are found in patients with HCM (85%) and RCM (8%) [5, 6] and rarely in DCM [7]. Interestingly, even within families, manifestations of different types of cardiomyopathy can be observed such as HCM and RCM that are caused by the same genetic defect. Contrary to HCM, RCM is characterised by restrictive filling and minimal or no left ventricular hypertrophy [8]. The presence of cardiomyocyte disarray in RCM, which is a histological hallmark of HCM, indicates that different types of cardiomyopathy fall within the same disease spectrum. Functional studies on RCM-causing TNNI3 mutations demonstrated a much higher increase in calcium sensitivity compared with HCM-causing mutations, which resulted in a more exaggerated HCM-like phenotype. Recently, TNNI3 mutations were also reported in autosomal dominant DCM [7]. Functional assays showed that these DCM causing TNNI3 mutations resulted in lower maximum ATPase rates and decreased calcium sensitivity compared with HCM [7]. These findings corroborate with previously described alterations in DCM caused by a mutation in the thin filaments.
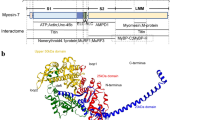
The majority of the TNNI3 mutations (85%) have been identified in exon 7 and exon 8, encoding domains interacting with cardiac actin (ACTC1) and cardiac troponin C (TNNC1) ([4]; Fig. 1). The positions of reported mutations in TNNI3 reveal the presence of important functional domains, such as protein binding or regulatory domains [9], such as phosphorylation sites in the N-terminal part of cardiac troponin [10].
Schematic representation of the TNNI3 gene, the corresponding protein structure and the sites of interaction with sarcomeric contractile proteins. Mutations identified in this study and majority known from literature are indicated. Mutations identified in RCM patients are indicated in red, mutations identified in HCM patients are indicated in black and mutations identified in DCM patients are indicated in blue. The (novel) RCM mutations found in this study are indicated in bold red. Boxes indicate exons, thin lines indicate introns and thick lines indicate interacting domains. Phosphorylation sites are indicated by PP
Mutations in TNNI3 have been reported in 2–7% of HCM cases [3, 11]. As literature data are often population specific, we investigated the prevalence of TNNI3 mutations within the Dutch cardiomyopathy population derived from four cardiogenetic centres. Based on the literature, the low frequency of TNNI3 cases might be explained by the severe clinical outcome of certain mutations. The fact that such rare mutations are still present in the population can be explained by the incidence of recurrent mutations due to mutational hot spots (codons with high mutation rate) or by founder mutations. In this study we combined patients and genetic data from four Dutch cardiogenetic centres, which offer diagnostic genetic testing of the TNNI3 gene in the entire country of the Netherlands.
Methods
Patient selection
Four Clinical Genetics Centres in the Netherlands (Rotterdam, Amsterdam, Maastricht and Groningen) offer genetic testing of the TNNI3 gene. All index patients underwent cardiological evaluation, consisting at least of a 12-lead ECG, transthoracic echocardiography and analysis of the family history. A total of 1040 index patients were included up to 2009. From Amsterdam we included 298 HCM patients, from Maastricht 250 HCM and 4 RCM patients, from Groningen 130 HCM and/or DCM patients and from Rotterdam 358 HCM index patients.
The postal codes of index patients carrying a mutation in TNNI3 were collected and converted into an overview of the Dutch postal code areas, based on the so called PC2 codes (the first two numbers of the postal code). These comprise 90 different Dutch regions, each containing on average 180,000 inhabitants.
Genetic evaluation
Total genomic DNA of the 1040 index patients (and 10 unaffected control persons) was extracted from EDTA (peripheral) blood samples according to standard procedures. Polymerase chain reaction (PCR) amplification and subsequent pre-screening using denaturing gradient gel electrophoresis (DGGE) and/or sequence analysis of the TNNI3 gene was performed using primers located in flanking intronic sequences (available on request). Nomenclature for the description of the mutation is in accordance with the Human Genome Variation Society (HGVS) using NM_000363.4 as the mRNA reference sequence. Microsatellite markers D19S402, D19S571, D19S921, D19S926, D19S880, D19S891, D19S210 and D19S887 flanking the TNNI3 gene were used for haplotype analysis (details about primers, PCR condition and fragment length are available upon request).
Results
The total number of patients with cardiomyopathy screened in this study was 1040. Among these patients, 14 different mutations (30 families in total) were found in TNNI3 indicating a mutation frequency of 3% in the Netherlands. Mean age at diagnosis was 36.5 years (3 months to 69 years, Table 1). All 14 mutations were detected in exon 7 or 8, and some mutations were found more frequently than others. Three of these mutations occurred more than four times in the Netherlands (see also Fig. 2 and Table 1); p.Arg145Trp (8 times), p.Ser166Phe (4 times) and p.Glu209Ala (5 times).
The p.Arg145Trp is the most frequent TNNI3 mutation in the Netherlands (27% of all TNNI3 mutations) and is randomly distributed over the country (Fig. 2). To investigate whether these families have a common ancestor, we performed haplotype analysis using eight polymorphic markers spanning the TNNI3 gene. Indeed, individuals carrying the mutation p.Arg145Trp show a unique short haplotype for markers D19S880, D19S891 and D19S210 within a region of 0.5 Mb (haplotype allele length respectively 96 bp, 177 bp and 105 bp). Unaffected controls or probands with another type of mutation lack this specific haplotype. Families carrying the p.Arg145Trp mutation mainly showed LVH and ECGs typical for HCM. Although not all ECG data were available from all individuals for our study (Table 1), it is interesting to see that all patients who had a clinical diagnosis of HCM by echocardiography also had ECG abnormalities. Most patients with a p.Arg145Trp mutation showed LVH and clear clinical symptoms between the age of 40 and 50 years. In three families with this mutation, cases of sudden death had occurred: in one family at age 41 (RCM, family W in Table 1) and in the other family (HCM, family AE in Table 1) at age 50. In the third family (HCM, family AD in Table 1) two persons died suddenly at ages 13 and 38 years respectively (no material was available to confirm the presence of the mutation), while in the same family the sister of the index patient with the mutation appeared to be unaffected at the age of 67. One proband (family Q) survived an out-of-hospital cardiac arrest. Differences in clinical expression were also observed in other families with the p.Arg145Trp mutation. It is not known whether this difference in clinical expression, ranging from very severe to unaffected, is due to an additional genetic mutation or environmental cause.
The p.Ser166Phe mutation was found in four index patients, three patients were from the same region within the province of Limburg. This is suggestive for a founder mutation and indeed, haplotype analysis showed that p.Ser166Phe carrying individuals also have a unique short haplotype for markers D19S880, D19S891 and D19S210 within a region of 0.5 Mb (haplotype allele length 100 bp, 187 bp and 99 bp, respectively). The amino acid Ser166 is highly conserved and located within the troponin C binding domain. In addition, Ser166 is known as one of the phosphorylation sites within TNNI3, involved in binding capacity to troponin C [12]. These observations and results from literature [3, 11] indicate that this is a pathogenic mutation. The time of disease onset also seems to be in the 4th decade of life, similar to patients having the p.Arg145Trp mutation.
The mutation p.Glu209Ala was found in five probands and is present in two different restricted areas, Groningen and Zeeland (Fig. 2). Interestingly, this mutation has not been described in the literature before. Unfortunately, the precise age of diagnosis is not known for all index patients, but also seems to be around the 4th decade of life. In four out of five patients haplotype analysis was performed and at least three different haplotypes surrounding the mutation were observed, indicating the presence of a recurring mutation due to a hot-spot rather than the presence of a founder mutation.
Three of four idiopathic RCM cases that carried a TNNI3 mutation were proven ‘de novo’ and already presented during childhood (ranging from birth to age of 10 years). RCM cases as indicated in Table 1 fulfilled RCM criteria such as impaired ventricular filling, normal or reduced diastolic filling, normal wall thickness and normal systolic function [13]. A de novo splice site mutation was detected in a young girl (age 6 months) with severe RCM, who died at the age of 1 year. One RCM-causing mutation, p.Arg145Trp, was not de novo. It was found in a 19 years-old index patient, and displayed clinical heterogeneity within the family (HCM and RCM) (family W, Table 1). The p.Arg145Trp is a founder mutation, which is found in our study mainly in combination with HCM.
Another novel RCM causing mutation p.Arg192Cys was found in combination with p.Arg145Gln in a young girl at age 9 years. The latter mutation was found in a mosaic status, which means that the p.Arg145Gln is not present in all cells (details not shown) and therefore p.Arg192Cys is probably the causative mutation. Unfortunately, the parents were not referred for genetic testing, so no definitive conclusions can be made regarding the origin of these two mutations.
Discussion
In this study we identified 14 different mutations in TNNI3, of which some mutations were found more frequently than others. A frequency of 3% TNNI3 mutation in Dutch cardiomyopathy families (1040 index patients) is in line with data from Mogensen et al. who studied 748 consecutive families. Most mutations in TNNI3 described up till now are found in exon 7 and 8, with some of these mutations found more frequently than others. Concordantly, we also identified most mutations in these exons.
The mutation Arg145Trp has frequently been reported in the literature and was also found in eight families in our study. The p.Arg145Trp mutation was identified in different regions of the Netherlands. In most families so far only the index patient has been investigated. Haplotype analysis showed in seven families (ten carriers in total) that the mutation arose from a common ancestor (founder effect). An explanation for the presence of founder mutations is that some mutations are better tolerated than others. Indeed, in the literature one large family (Arg145Trp) was described with more than four gene carriers who showed late onset and low-risk disease without progression during follow-up over 3 years [14–16]. None of the family members in this large family had any risk factors for sudden death. The proband had the most prominent LVH associated with outflow tract obstruction. Two other individuals fulfilled criteria for familial HCM and had abnormal ECGs but normal echocardiograms. Magnetic resonance imaging (MRI) detected LVH in both individuals. However, our study shows that two out of eight founder carriers have disease onset at a young age (age of 19 (RCM) and one even before age of 9). A family member of the 19-year-old RCM patient died suddenly at age 41. The difference in disease expression and disease onset for this founder mutation (even within one family, see for example family AD in Table 1) might be explained by additional mutations present in other known or unknown genes encoding for sarcomeric, cytoskeleton or nuclear envelope proteins next to the mutation found in TNNI3.
In addition to the Arg145Trp mutation, Arg145Gly and Arg145Gln [17] mutations have been identified affecting the same amino acid position. In one of our patients (19 years old) with RCM we found the founder p.Arg145Trp substitution, which was also reported for two other cases of RCM, who surprisingly had a relatively old age (67 and 70 years) at first presentation [8]. Since different mutations affect the same codon, we speculate that this codon 145 is also subject to a high mutation rate, which can lead to new patients having either RCM or HCM,
The mutation p.Ser166Phe was found four times and previously reported in the literature [11, 14]. Haplotype analysis indicated that the p.Ser166Phe is a founder mutation in the Netherlands. Almost all affected carriers in our study and those known from literature [11] have obstructive HCM (in some cases myectomy was performed) including asymmetrical septum hypertrophy and ECG abnormalities. The mutation affects a PKA phosphorylation site [12] and phosphorylation of this site influences the interaction of troponin I with the calcium-binding site of troponin C [9]. HCM-causing missense mutations within the amphipathic alpha-helix of TNNI3, spanning amino acids 164 to 188 and probably involved in the binding to actin-tropomyosin, cause an increased calcium sensitivity while DCM-causing mutations cause a decreased sensitivity [7].
The p.Glu209Ala has never been reported before, but in the Netherlands it has been found in five index patients so far. According to genetic criteria, it would have been labelled as an unclassified variant of unknown pathological significance (UV). However, the fact that this recurrent mutation has been detected multiple times independently in different centres supports a pathogenic character. The position of the amino acid (p.Glu209), residing one codon from the C-terminus end, may indicate an important biological function. Several experimental studies have indicated that residues located within the C terminus of troponin I are involved in interaction with the regulatory domain (N-terminal domain) of troponin C [18]. The C-terminus of troponin I is highly conserved among the troponin isoforms and its binding to the N terminus of troponin C, which contains the low affinity calcium-specific site, indicates that it may be important for calcium-dependent regulation of cardiac muscle contraction [19].
A relatively high incidence of de novo TNNI3 mutations was detected in young children (especially in young females with RCM (2 out of 4)). The absence of family history but presence of RCM in an index patient is a strong indication for diagnostic TNNI3 mutation detection. This is similar to the results by Mogensen [3], who reported that 30% of TNNI3 mutations in RCM are caused by a de novo mutation. Until now, all mutations found in TNNI3 are missense mutations (Fig. 1). A splice site mutation in TNNI3 causing RCM has never been reported before and probably causes a truncated protein with a severe dominant negative effect. So the extreme severity of the disease in this family was probably due to the underlying molecular effect.
Conclusion
TNNI3 mutations have been identified in 3% of the Dutch cardiomyopathy patients, with a relatively high amount of frequently occurring mutations. Different mutations that occur at the same position can lead to different forms of cardiomyopathy with variable penetrance. Our findings show that disease-causing mutations can lead to either HCM, RCM or DCM (or a combination of both within families) depending on the specific mutation. We hypothesise that the presence of RCM and other cardiomyopathies within one family might still be explained by additional mutations present in genes encoding cytoskeleton or nuclear envelope proteins additional to the TNNI3 mutation. Even in families with a proven ‘founder mutation’, the phenotypic heterogeneity is remarkable, indicating that other genetic background cannot be ruled out and might be explained by linked or even unlinked genetic modifiers. In each case of RCM, especially in young patients, TNNI3 analysis should still be the initial gene to be excluded.
References
Alcalai R, Seidman JG, Seidman CE. Genetic basis of hypertrophic cardiomyopathy: from bench to the clinics. J Cardiovasc Electrophysiol. 2008;19(1):104–10. Epub 2007 Oct 4.
Gomes AV, Potter JD. Molecular and cellular aspects of troponin cardiomyopathies. Ann NY Acad Sci. 2004;1015:214–24.
Mogensen J, Murphy RT, Kubo T, et al. Frequency and clinical expression of cardiac troponin I mutations in 748 consecutive families with hypertrophic cardiomyopathy. J Am Coll Cardiol. 2004;44(12):2315–25.
Kimura A, Harada H, Park JE, et al. Mutations in the cardiac troponin I gene associated with hypertrophic cardiomyopathy. Nat Genet. 1997;16(4):379–82.
Doolan A, Tebo M, Ingles J, et al. Cardiac troponin I mutations in Australian families with hypertrophic cardiomyopathy: clinical, genetic and functional consequences. J Mol Cell Cardiol. 2005;38(2):387–93.
Kaski JP, Syrris P, Burch M, et al. Idiopathic restrictive cardiomyopathy in children is caused by mutations in cardiac sarcomere protein genes. Heart. 2008;94(11):1478–84.
Carballo S, Robinson P, Otway R, et al. Identification and functional characterization of cardiac troponin I as a novel disease gene in autosomal dominant dilated cardiomyopathy. Circ Res. 2009;105(4):375–82.
Kubo T, Gimeno JR, Bahl A, et al. Prevalence, clinical significance, and genetic basis of hypertrophic cardiomyopathy with restrictive phenotype. J Am Coll Cardiol. 2007;49(25):2419–26. Epub 2007 Jun 11.
Solaro RJ, Rosevear P, Kobayashi T. The unique functions of cardiac troponin I in the control of cardiac muscle contraction and relaxation. Biochem Biophys Res Commun. 2008;369(1):82–7.
Howarth JW, Meller J, Solaro RJ, Trewhella J, Rosevear PR. Phosphorylation-dependent conformational transition of the cardiac specific N-extension of troponin I in cardiac troponin. J Mol Biol. 2007;373(3):706–22.
Van Driest SL, Ellsworth EG, Ommen SR, Tajik AJ, Gersh BJ, Ackerman MJ. Prevalence and spectrum of thin filament mutations in an outpatient referral population with hypertrophic cardiomyopathy. Circulation. 2003;108(4):445–51.
Ward DG, Ashton PR, Trayer HR, Trayer IP. Additional PKA phosphorylation sites in human cardiac troponin I. Eur J Biochem. 2001;268(1):179–85.
Kushwaha SS, Fallon JT, Fuster V. Restrictive cardiomyopathy. N Engl J Med. 1997;336(4):267–76.
Mogensen J, Kubo T, Duque M, et al. Idiopathic restrictive cardiomyopathy is part of the clinical expression of cardiac troponin I mutations. J Clin Invest. 2003;111(2):209–16.
Fokstuen S, Lyle R, Munoz A, et al. A DNA resequencing array for pathogenic mutation detection in hypertrophic cardiomyopathy. Hum Mutat. 2008;29(6):879–85.
Andersen PS, Havndrup O, Hougs L, et al. Diagnostic yield, interpretation, and clinical utility of mutation screening of sarcomere encoding genes in Danish hypertrophic cardiomyopathy patients and relatives. Hum Mutat. 2009;30(3):363–70.
Moon JC, Mogensen J, Elliott PM, et al. Myocardial late gadolinium enhancement cardiovascular magnetic resonance in hypertrophic cardiomyopathy caused by mutations in troponin I. Heart. 2005;91(8):1036–40.
Tripet B, Van Eyk JE, Hodges RS. Mapping of a second actin-tropomyosin and a second troponin C binding site within the C terminus of troponin I, and their importance in the Ca2+−dependent regulation of muscle contraction. J Mol Biol. 1997;271(5):728–50.
Rarick HM, Tu XH, Solaro RJ, Martin AF. The C terminus of cardiac troponin I is essential for full inhibitory activity and Ca2+ sensitivity of rat myofibrils. J Biol Chem. 1997;272(43):26887–92.
Acknowledgements
Francis van der Lubbe for making TNNI3 Fig. 1.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
van den Wijngaard, A., Volders, P., Van Tintelen, J.P. et al. Recurrent and founder mutations in the Netherlands: cardiac Troponin I (TNNI3) gene mutations as a cause of severe forms of hypertrophic and restrictive cardiomyopathy. Neth Heart J 19, 344–351 (2011). https://doi.org/10.1007/s12471-011-0135-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12471-011-0135-z