Abstract
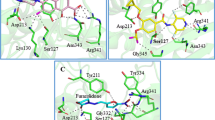
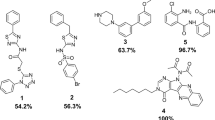
The gram-positive bacterium Staphylococcus aureus, responsible for a wide variety of diseases in human involve all organ systems ranging from localized skin infections to life-threatening systemic infections. FtsZ, the key protein of bacterial cell division was selected as a potent anti bacterial target. In order to identify the new compounds structure based screening process was carried out. An enrichment study was performed to select a suitable scoring function and to retrieve potential candidates against FtsZ from a large chemical database. The docking score and docking energy values were compared and their atomic interaction was also evaluated. Furthermore molecular dynamics simulation were also been performed to check the stability and the amino acids interacted towards the FtsZ. Finally we selected C ID 16284, 25916, 15894, 13403 as better lead compounds. From these results, we conclude that our insilico results will provide a framework for the detailed in vitro and in vivo studies about the FtsZ protein activity in drug development process.
Similar content being viewed by others
References
Anne Marie, J., Sid T. 2008. Driving forces for ligand migration in the leucine transporter. Chem Biol Drug Des 72, 265–272.
Bowers, K.J., Chow, E., Xu H., Dror R.O., Eastwood M.P., Gregersen B.A., Klepeis J.L., Kolossvary I., Moraes M.A., Sacerdoti F.D., Salmon J.K., Shan Y., Shaw D.E. 2006. Scalable algorithms for molecular dynamics simulations on commodity clusters. ACM, New York, NY 84: 1145/1188455.1188544.
Bowie, J.U., Luthy, R., Eisenberg, D. 1991. A method to identify protein sequences that fold into a known three-dimensional structure. Science 253, 164–170.
Bramhill, D. 1997. Bacterial cell division. Ann Rev Cell & Dev Biol 13, 395–424.
Clark, D.E. 2008. What has virtual screening ever done for drug discovery? Expert opin drug dis 3, 841–851.
Dai, K., Mukherjee, A., Xu, Y., Lutkenhaus, J. 1994. Mutations in FtsZ that confer resistance to SulA affect the interaction of FtsZ with GTP. J Bacteriol 176, 130–136.
Dasgupta, D. 2009. Novel compound with potential of an antibacterial drug targets FtsZ protein. Biochem J 423, e1–e3.
De Boer, P., Crossley, R.E., Rothfield, L.I. 1988. Isolation and properties of minB, a complex geneticlocus involved in correct placement of the division site in Escherichia coli. J Bacteriol 170, 2106–2112.
Duffy, E.M., Jorgensen, W.L, 2000. Prediction of properties from simulations: free energies of solvation in hexadecane, octanol, and water. J Am Chem Soc 122, 2878–2888.
Fernandez-Recio, J., Totrov, M., Skorodumov, C., Abagyan, R. 2005. Optimal docking area: a new method for predicting protein-protein interaction sites. Proteins 58(1), 134–143.
Foster, T.J., Hook, M. 1998. Surface protein adhesins of Staphylococcus aureus. Trends Microbiol 6, 484–488.
Friesner, R.A., Banks, J.L., Murphy, R.B., Halgren, T.A., Klicic, J.J., Mainz, D.T., Repasky, M.P., Knoll, E.H., Shelley, M., Perry, J.K. 2004. Glide: a new approach for rapid, accurate docking and scoring: method and assessment of docking accuracy. J Med Chem 47, 1739–1749.
Friesner, R.A., Banks, J.L., Murphy, R.B., Halgren, T.A., Klicic, J.J., Mainz, D.T., Repasky, M.P., Knoll, E.H., Shelley, M., Perry, J.K., Shaw, D.E., Francis, P., Shenkin, P.S. 2004. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem 47, 1739–1749.
Friesner, R.A., Murphy, R.B., Repasky, M.P., Frye, L.L., Greenwood, J.R., Halgren, T.A., Sanschagrin, P.C., Mainz, D.T. 2006. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein-ligand complexes. J Med Chem 49(21), 6177–6196.
Galeazzi, Roberta. 2009. Molecular Dynamics as a Tool in Rational Drug Design: Current Status and Some Major Applications. Curr Comput Aided Drug Des 4, 225–240.
Gordon, R.J., Lowy, F.D. 2008. Pathogenesis of methicillin-resistant Staphylococcus aureus infection. Clin Infect Dis 46, 5S350–S359.
Gurung, M., Chan Moon, D., Chai, C.W., Lee, J.H., Bae, Y.C., Kim, J., Lee, Y.C., Seol, S.Y., Cho, D.T., Kim, S., Lee, J.C. 2011. Staphylococcus aureus Produces Membrane-Derived Vesicles That Induce Host Cell Death. Plos one 6: e27958.
Halgren, T. 2007. New method for fast and accurate binding-site identification and analysis. Chem Biol Drug Des 69(2), 146–148.
Halgren, T.A. 2009. Identifying and characterizing binding sites and assessing druggability. J Chem Inf Model 49(2), 377–389.
Jorgensen, W.L. 2004. The Many Roles of Computation in Drug Discovery. Science 303, 1813–1818.
Keiichi, H. 2001. Vancomycin-resistant Staphylococcus aureus: a new model of antibiotic resistance. Lancet Infect Dis 1, 147–155.
Leach, A.R., Shoichet, B.K., Peishoff, C.E. 2006. Prediction of Protein — Ligand Interactions Docking and scoring: Successes and Gaps. J Med Chem 49, 5851–5855.
Li, L., Darden, T., Hiskey, R., Pedersen, L. 1996. Homology modeling and molecular dynamics simulations of the Gla domains of human coagulation factor IX and Its G [12]A mutant. J Phys Chem 100, 2475–2479.
Lipinski, C.A., Lombardo, F., Dominy, B.W., Feeney, P.J. 1997. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 23, 3–25.
Lock, R.L., Harry, E.J. 2008. Cell-division inhibitors: new insights for future antibiotics. Nat Rev Drug Discov 7, 324–338.
Lyne, P.D., Lamb, M.L., Saeh, J.C 2006. Accurate prediction of the relative potencies of members of a series of kinase inhibitors using molecular docking and MMGBSA scoring. J Med Chem 49, 4805–4808.
Manjasetty, B.A., Turnbull, A.P., Panjikar, S., Bussow, K., Chance, M.R. 2008. Automated technologies and novel techniques to accelerate protein crystallography for structural genomics. Proteomics 8, 612–625.
Maragakis, P., Lindorff-Larsen, K., Eastwood, M.P., Dror, R.O., Klepeis, J.L., Arkin, I.T., Jensen, M. Xu, H., Trbovic, N., Friesner, R.A., Iii, A.G., Shaw, D.E. 2008. Microsecond molecular dynamics simulation shows effect of slow loop dynamics on backbone amide order parameters of proteins. J Phys Chem B 112(19), 6155–6158.
Mukherjee, A., Lutkenhaus, J. 1994. Guanine nucleotide-dependent assembly of FtsZ into filaments. J Bacteriol 176, 2754–27558.
Ohtsuki, S., Uchida, Y., Kubo, Y., Terasaki, T. 2011. Quantitative Targeted Absolute Proteomics-Based ADME Research as a New Path to Drug Discovery and development: methodology, advantages, strategy and prospects. J Pharm Sci 100, 3547–3559.
Prashantha Kumar B.R., Soni, M., Bharvi Bhikhalal, U., Ismayil, R.K., Jagadeesh, M., Bommu, P., Nanjan, M.J. 2010. Analysis of physicochemical properties for drugs from nature. Med Chem Res 19, 984–992.
Ray, Chaudhuri, D., Park, J.T. 1992. Escherichia coli cell-division gene ftsZ encodes a novel GTP-binding protein. Nature 359, 251–254.
Selvaraj, C., Singh, S.K., Tripathi, S.K., Reddy, K.K., Rama, M. 2011. In silico screening of indinavir-based compounds targeting proteolytic activity in HIV PR: binding pocket fit approach. Medicinal Chemistry Research l 1–19.
Shopsin, B., Mathema, B., Martinez, J., Ha, E., Campo, M.L., Fierman, A., Krasinski, K., Kornblum, J., Alcabes, P., Waddington, M., et al 2000. Prevalence of methicillin-resistant and methicillin susceptible Staphylococcus aureus in the community. J Infect Dis 182, 359–362.
Singh, V., Somvanshi, P. 2009. Homology modelling of 3-oxoacyl-acyl carrier protein synthase II from Mycobacterium tuberculosis H37Rv and molecular docking for exploration of drugs. J Mol Model 15, 453–460.
Sitemap, version 2.4 (2008) Schrödinger, LLC: New York, NY
Vollmer, W. 2006. The prokaryotic cytoskeleton: a putative target for inhibitors and antibiotics. ApplMicrobiol Biotechnol 73, 37–47.
Wu, X., Ohrngren, P., Ekegren, J.K., Unge, J., Unge, T., Wallberg, H., Samuelsson, B., Hallberg, A., Larhed, M. 2008. Two-carbonelongated HIV-1 protease inhibitors with a tertiary-alcoholcontaining transition-state mimic. J Med Chem 51, 1053–1057.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Vijayalakshmi, P., Nisha, J. & Rajalakshmi, M. Virtual screening of potential inhibitor against FtsZ protein from Staphylococcus aureus . Interdiscip Sci Comput Life Sci 6, 331–339 (2014). https://doi.org/10.1007/s12539-012-0229-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12539-012-0229-3