Abstract
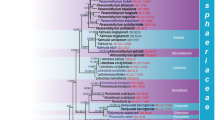
The phylogenetic relationships of Lasiosphaeriaceae are complicated in that the family is paraphyletic and includes Sordariaceae and Chaetomiaceae, as well as several polyphyletic genera. This study focuses on the phylogenetic relationships of the coprophilous genera, Anopodium, Apodospora, Arnium, Fimetariella and Zygospermella. They are traditionally circumscribed based on ascospore characters, which have proven homoplasious in other genera within the family. Our results based on LSU nrDNA and ß–tubulin sequences distinguish four lineages of Lasiosphaeriaceae taxa. Anopodium joins the clade of morphologically similar, yellow-pigmented species of Cercophora and Lasiosphaeria. Apodospora is monophyletic and joins a larger group of taxa with unclear affinities to each other, while Arnium is polyphyletic being scattered throughout three of the four major clades of Lasiosphaeriaceae. Fimitariella is represented by a single collection and joins the clade containing Cercophora scortea and Podospora appendiculata. Zygospermella shows affinities to the Lasiosphaeris clade. Based on a combination of morphological and molecular data, Echria stat. nov. is recognized at the genus level for the former Arnium section and two new combinations are proposed: E. gigantospora and E. macrotheca.




Similar content being viewed by others
References
Alexopoulos CJ, Mims CW, Blackwell M (1996) Introductory mycology. Wiley, New York
Alfaro ME, Zoller S, Lutzoni F (2003) Bayes or bootstrap? A simulation study comparing the performance of Bayesian Markov Chain Monte Carlo sampling and bootstrapping in assessing phylogenetic confidence. Mol Biol Evol 20:255–266
von Arx JA, Guarro J, Figueras MJ (1986) The ascomycete genus Chaetomium. Beih Nova Hedwig 84:1–162
Bell A, Mahoney D, Debuchy R (2008) A new record of Apodospora from Australia, a rarely collected coprophilous ascomycete. Aust Mycol 27:136–140
Cai L, Jeewon R, Hyde KD (2006) Molecular systematics of Zopfiella and allied genera: evidence from multi-gene sequence analysis. Mycol Res 110:359–368
Cain RF, Mirza JH (1970) Apodospora, a new genus of the Sordariaceae. Can J Bot 48:891–896
Cannon PF, Kirk PM (2007) Fungal families of the world. CABI, Wallingford
Chang J-H, Kao H-W, Wang Y-Z (2010) Molecular phylogeny of Cercophora, Podospora, Schizothecium (Lasiosphaeriaceae, Pyrenomycetes). Taiwania 55:110–116
Dettman JR, Harbinski FM, Taylor JW (2001) Ascospore morphology is a poor predictor of the phylogenetic relationships of Neurospora and Gelasinospora. Fungal Genet Biol 34:49–61
Furuya K, Udagawa S (1976) Coprophilous pyrenomycetes from Japan IV. T Mycol Soc Jpn 17:248–261
Garcia D, Stchigel AM, Cano J, Guarro J, Hawksworth DL (2004) A synopsis and re-circumscription of Neurospora (syn. Gelasinospora) based on ultrastructural and 28S rDNA sequence data. Mycol Res 108:1119–1142
Greif MD, Stchigel AM, Miller AN, Huhndorf SM (2009) A re-evaluation of genus Chaetomidium based on molecular and morphological characters. Mycologia 101:554–564
Hilber R, Hilber O (1979) Einige Anmerkungen zu der Gattung Cercophora Fuckel (Lasiosphaeriaceae). Z Mykol 45:209–233
Hillis DM, Bull JJ (1993) An empirical test of bootstrapping as a method for assessing confidence in phylogenetic analysis. Syst Biol 42:182–192
Huhndorf SM, Miller AM, Fernández FA (2004) Molecular systematics of the Sordariales: the order and the family Lasiosphaeriaceae redefined. Mycologia 96:368–387
Huhndorf SM, Miller AM (2011) A molecular re-appraisal of taxa in the Sordariomycetidae and a new species of Rimaconus from New Zealand. Stud Mycol 68:203–210
Katoh K, Misawa K, Kuma K, Miyata T (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066
Krug JC (1995) The genus Fimetariella. Can J Bot 73:1905–1916
Krug JC, Cain RF (1972) Additions to the genus Arnium. Can J Bot 50:367–373
Lanfear R, Calcott B, Ho S, Guindon S (2012) PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol 29:1695–1701
Lundqvist N (1964a) Anopodium, a new genus of coprophilous Pyrenomycetes with apically pedicellate spores. Bot Notiser 117:355–365
Lundqvist N (1964b) Fimetariella, a new genus of coprophilous Pyrenomycetes. Bot Notiser 117:238–248
Lundqvist N (1967) On spore ornamentation in the Sordariaceae, exemplified by the new cleistocarpous genus Copromyces. Ark Bot, Ser 2, 6:327–337
Lundqvist N (1969) Zygopleurage and Zygospermella (Sordariaceae s. lat., Pyrenomycetes). Bot Notiser 122:353–374
Lundqvist N (1972) Nordic Sordariaceae s. lat. Symb Bot Ups 20:1–374
Mason-Gamer RJ, Kellogg EA (1996) Testing for phylogenetic conflict among molecular data sets in the tribe Triticeae (Gramineae). Syst Biol 45:524–545
Miller AN, Huhndorf SM (2004) A natural classification of Lasiosphaeria based on nuclear LSU rDNA sequences. Mycol Res 108:26–34
Miller AN, Huhndorf SM (2005) Multi-gene phylogenies indicate ascomal wall morphology is a better predictor of phylogenetic relationships than ascospore morphology in the Sordariales (Ascomycota, Fungi). Mol Phylogenet Evol 35:60–75
Mirza JH, Cain RF (1969) Revision of the genus Podospora. Can J Bot 47:1999–2048
de Queiroz A (1993) For consensus (sometimes). Syst Biol 42:368–372
Raja HA, Shearer CA (2006) Arnium gigantosporum, a new ascomycete species from fresh water in Florida. Fungal Divers 22:219–225
Rambaut A, Drummond AJ (2009) Tracer v1.5.0. Available from http://beast.bio.ed.ac.uk
Réblová M (2008) Bellojisia, a new sordariaceous genus for Jobellisia rhynchostoma and a description of Jobellisiaceae fam. nov. Mycologia 100:893–901
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Stamatakis A (2006) RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690
Stamatakis A, Hoover P, Rougemont J (2008) A Rapid Bootstrap Algorithm for the RAxML Web Servers. Syst Biol 57:758–771
Stchigel AM, Cano J, Miller AN, Calduch M, Guarro J (2006) Corylomyces: a new genus of Sordariales from plant debris in France. Mycol Res 110:1361–1368
Vilgalys R. Conserved primer sequences for PCR amplification and sequencing from nuclear ribosomal RNA. Duke University, USA. Internet address: http://www.biology.duke.edu/fungi/mycolab/primers.htm
Zhang N, Castlebury LA, Miller AN, Huhndorf SM, Schoch CL, Seifert KA, Rossman AY, Rogers JD, Kohlmeyer J, Volkmann-Kohlmeyer B, Sung GH (2006) An overview of the systematics of the Sordariomycetes based on a four-gene phylogeny. Mycologia 98:1076–1087
Acknowledgment
This study is financed by the Swedish Taxonomy Initiative (dha 34/07 1.4) and the Helge Ax:son Johnson foundation. This work was supported in part by a NSF PEET Grant (DEB–9,521,926) to SMH and some sequences were generated in the Pritzker Laboratory for Molecular Systematics and Evolution at The Field Museum of Natural History.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kruys, Å., Huhndorf, S.M. & Miller, A.N. Coprophilous contributions to the phylogeny of Lasiosphaeriaceae and allied taxa within Sordariales (Ascomycota, Fungi). Fungal Diversity 70, 101–113 (2015). https://doi.org/10.1007/s13225-014-0296-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13225-014-0296-3