Abstract
Background
Salt-tolerant breeding of maize has great significance to the development and utilization of saline–alkaline soil and the maintenance of grain security. Genome-wide association study (GWAS) has been widely used in maize genetics and breeding.
Objective
To discover new salt-tolerant genes in maize by association analysis, which can provide technical supports for the innovation and genetic improvement of salt-tolerant germplasm resources in maize.
Methods
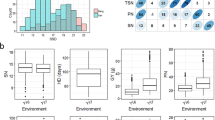
Totally 150 maize inbred lines were genotyped with a high-density chip. GWAS was carried out to identify the significant single nucleotide polymorphisms (SNPs) which were associated with maize salt tolerance. Totally 34,972 SNPs with high quality and diversity were selected from 56,110 SNP markers, which were distributed on 10 chromosomes of maize. The GLM algorithm in TASSEL5.2 was used to analyze the five traits related to salt tolerance.
Results
Using a strict LOD threshold of 4.5, totally 7 SNP loci were identified, which were significantly correlated with plant height change rate and fresh weight change rate. The high density fingerprints of 150 inbred lines were clustered by TASSEL5.2 software to construct genetic clustering map to estimate the genetic distance and the subgroups. The 150 maize inbred lines were divided into two groups: SS group and NSS group, and the SNP loci of the salt-tolerant index showed difference in chromosome distribution. Based on previous studies, we screened 8 candidate genes for salt tolerance in maize and four of them were further validated by real-time quantitative PCR.
Conclusion
Totally 7 SNP loci and 8 candidate genes related to salt tolerance in maize were identified, which will be of special value in molecular breeding of salt-tolerant maize.







Similar content being viewed by others
References
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23(19):2633–2635
Committee IAP (2014) USDA agricultural projections to 2023. United States Department of Agriculture
Deng M, Zou X, Huo J, Wen J, Zhu H (2010) Clone and sequence analysis of the specific mitochondrial genes cox and atp 6 related to cytoplasmic male sterile in pepper. Acta Bot Bor-Occid Sin 30(10):1935–1940
Di H, Liu X, Wang Q, Weng J, Zhang L (2015) Development of SNP-based dCAPS markers linked to major head smut resistance quantitative trait locus qHS2.09 in maize. Euphytica 202(1):69–79
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4(2):359–361
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14(8):2611–2620
Ganal M, Gregor D, Andreas P (2011) A large maize (Zea mays L.) SNP genotyping array: development and germplasm genotyping, and genetic mapping to compare with the B73 reference genome. PLoS ONE 6(12):1–15
Gore MA, Chia JM, Elshire RJ, Sun Q, Ersoz ES, Hurwitz BL, Peiffer JA, McMullen MD, Grills GS, Ross-Ibarra J (2009) A first-generation haplotype map of maize. Science 326(5956):1115–1117
Grebenok RJ, Galbraith DW, Penna DD (1997) Characterization of zea mays endosperm C-24 sterol methyltransferase: one of two types of sterol methyltransferase in higher plants. Plant Mol Biol 34:891–896
Lai J, Li R, Xu X, Jin W, Xu M, Zhao H, Xiang Z, Song W, Ying K, Zhang M (2010) Genome-wide patterns of genetic variation among elite maize inbred lines. Nat Genet 42(11):1027-U1158
Liu CL, Weng JF, Zhang DG, Zhang XC, Yang XY, Shi LY, Meng QC, Yuan JH, Guo XP, Hao ZF (2014a) Genome-wide association study of resistance to rough dwarf disease in maize. Eur J Plant Pathol 139(1):205–216
Liu Y, Wu H, Kou L, Liu X, Zhang J, Guo Y, Ma E (2014b) Two metallothionein genes in oxya chinensis: molecular characteristics, expression patterns and roles in heavy metal stress. PLoS One 9(11):e112759
Liu N, Xue Y, Guo Z (2016) Genome-wide association study identifies candidate genes for starch content regulation in maize kernels. Front Plant Sci 7(e28334):1–8
Luo M, Zhao Y, Zhang R (2017) Mapping of a major QTL for salt tolerance of mature field-grown maize plants based on SNP markers. BMC Plant Biol 17(1):140–150
Martinez-Beltran J, Licona Manzur C (2005) Overview of salinity problems in the world and FAO strategies to address the problem. In: Proceedings of the international salinity forum; Riverside, California, pp 311–314
Metzker ML (2010) Sequencing technologies - the next generation. Nat Rev Genet 11(1):31–46
Miyama M, Tada Y (2008) Transcriptional and physiological study of the response of Burma mangrove (Bruguiera gymnorhiza) to salt and osmotic stress. Plant Mol Biol 68(1–2):119–129
Mo J, Li D, Zhang H, Song F (2011) Roles of ERF transcription factors in biotic and abiotic stress response in plants. Acta Phytophysiol Sin 47(12):1145–1154
Ren F (2014) Evaluation of maize inbred lines germination characteristics and SNP association analysis. Master’s Dissertation, Shandong Agricultural University
Revilla P, Rodríguez Víctor Manuel, Ordás Amando, Rincent Renaud, Charcosset Alain, Giauffret Catherine, Melchinger Albrecht E, Schön Chris-Carolin, Bauer Eva, Altmann Thomas (2016) Association mapping for cold tolerance in two large maize inbred panels. BMC Plant Biol 16(1):127–137
Schnable P (2012) The B73 maize genome: complexity, diversity, and dynamics. Science 337(6098):1112–1115
Song W, Liu M (2017) Farmland conversion decreases regional and national land quality in china. Land Degrad Dev 28(2):459–471
Sun Y, Niu L, Song F (2014) Progress on salinization soil restoration method. Inter J Ecol 03(02):30–36
Tian F, Bradbury PJ, Brown PJ, Hung H, Sun Q, Flint-Garcia S, Rocheford TR, McMullen MD, Holland JB, Buckler ES (2011) Genome-wide association study of leaf architecture in the maize nested association mapping population. Nat Genet 43(2):159–162
Tong W, Chen X, Wen X, Chen F, Zhang H, Chu Q, Dikgwatlhe Batsile S (2015) Applying a salinity response function and zoning saline land for three field crops: a case study in the Hetao Irrigation District, Inner Mongolia, China. J Integr Agric 14(1):178–189
Unterseer S, Bauer E, Haberer G (2014) A powerful tool for genome analysis in maize: development and evaluation of the high density 600 k SNP genotyping array. BMC Genom 15(1):823–838
Wang X, Liu HWS, Ferjani A, Li J, Yan J, Yang X, Qin F (2016) Genetic variation in ZmVPP1 contributes to drought tolerance in maize seedlings. Nat Genet 48(10):1233–1241
Yang Z, Wang B (2014) Progress in techniques of improvement and utilization of saline-alkali land in china and its future trend. J Soil Water Conserv 02(01):1–11
Zhang M, Liang C (2000) Progress in the studies of lipid transfer protein. Prog Biochem Biophys 3(27):244–247
Zhang L, Niu X, Zhang Y, Liu Y (2012) Functional analysis via overexpressing xyloglucan endotransglycosylase gene OsXTH11 in rice. Sci Agric Sin 45(16):3231–3239
Zhang C, Jin Y, Liu J, Tang Y, Cao S, Qi H (2014) The phylogeny and expression profiles of the lipoxygenase (LOX) family genes in the melon (Cucumis melo L.) genome. Sci Hortic 170:94–102
Zuo W, Chao Q, Zhang N, Ye J, Tan G, Li B, Xing Y, Zhang B, Liu H, Fengler KA (2015) A maize wall-associated kinase confers quantitative resistance to head smut. Nat Genet 47(2):151–157
Acknowledgements
This work was supported by the Key Research and Development Project of Jiangsu Province, China (Modern Agriculture, BE2017365), the State Key Laboratory of Crop Biology (Shandong Agricultural University) Open Fund (2016KF02), the fund from Nantong Xinhe Bio-Technology Co. Ltd. (17ZH050), and the Practice Innovation Training Program Projects for College Students.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Xie, Y., Feng, Y., Chen, Q. et al. Genome-wide association analysis of salt tolerance QTLs with SNP markers in maize (Zea mays L.). Genes Genom 41, 1135–1145 (2019). https://doi.org/10.1007/s13258-019-00842-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-019-00842-6