Abstract
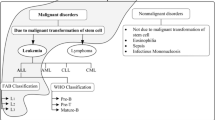
Microscopic pathology is still a meticulous and biased task for hematologist, which leads to the misclassification of cells and vagueness prediction of abnormal cells due to variability in the morphological structure of leukocytes. Therefore, to enhance the detection precision and diminishing the time factor, an automatic classification system for leukocytes has been proposed. In routine clinical practice, expert hematologists observed that the nucleus plays a crucial role in the identification of the blood disorders. Accordingly, in this work, the localization of leukocyte nucleus is performed by using Chan–Vase level-set method for the design of a classification framework that differentiates between four classes of the leukocytes, i.e., eosinophils, polymorphs, monocytes and lymphocytes based on the nucleus. A dataset consisting of 162 leukocyte microscopic images is used. The images in the dataset are classified on the basis of texture, shape and color features. The feature selection method based on the linguistic hedge is applied on evaluated feature space of 92. The selected features are fed to an adaptive neuro-fuzzy classifier for the classification. The proposed framework obtained an accuracy of 98.7% after applying the adaptive neuro-fuzzy classification on selected 46 informative features. The correlation of best features and data extorted from the different microscopic images may yield a dramatic increase in diagnostic consistency in clinical pathology. The results obtained by utilization of selected optimal features and adaptive neuro-fuzzy classification system indicate that it can be routinely used in clinical environment for differential diagnosis between different classes of leukocytes.
Similar content being viewed by others
References
Putzu, L.: Computer aided diagnosis algorithms for digital microscopy. Doctoral dissertation, Universita’degli Studi di Cagliari.
Putzu, L.; Caocci, G.; Di Ruberto, C.: Leucocyte classification for leukemia detection using image processing techniques. Artif. Intell. Med. 62(3), 179–91 (2014)
Bhattacharjee, S.; Mukherjee, J.; Nag, S.; Maitra, I.K.; Bandyopadhyay, S.K.: Review on histopathological slide analysis using digital microscopy. Int. J. Adv. Sci. Technol. 62, 65–96 (2014)
Yu, W.; Lü, J.; Yu, X.; Chen, G.: Distributed adaptive control for synchronization in directed complex networks. SIAM J. Control Optim. 53(5), 2980–3005 (2015)
Liu, K.; Wu, L.; Lü, J.; Zhu, H.: Finite-time adaptive consensus of a class of multi-agent systems. Sci. China Technol. Sci. 59(1), 22–32 (2016)
Kumar, I.; Bhadauria, H.S.; Virmani, J.; Thakur, S.: A hybrid hierarchical framework for classification of breast density using digitized film screen mammograms. Multimed. Tools Appl. 76, 18789–18813 (2017)
Pang, G.; Zhuang, Y.; Zhou, P.: Automatic leukocytes classification by distance transform, moment invariant, morphological features, gray level co-occurrence matrices and SVM. In: International Conference on Information Sciences, Machinery, Materials and Energy (ICISMME 2015), pp. 1090–1095
Nazlibilek, S.; Karacor, D.; Ertürk, K.L.; Sengul, G.; Ercan, T.; Aliew, F.: White blood cells classifications by SURF image matching. PCA and dendrogram. Biomed. Res. 26(4), 633–640 (2015)
Ravikumar, S.; Shanmugam, A.: WBC image segmentation and classification using RVM. Appl. Math. Sci. 8(45), 2227–37 (2014)
Nazlibilek, S.; Karacor, D.; Ercan, T.; Sazli, M.H.; Kalender, O.; Ege, Y.: Automatic segmentation, counting, size determination and classification of white blood cells. Measurement 30(55), 58–65 (2014)
Habibzadeh, M.; Krzyżak, A.; Fevens, T.: Comparative study of shape, intensity and texture features and support vector machine for white blood cell classification. J. Theor. Appl. Comput. Sci. 7(1), 20–35 (2013)
Rezatofighi, S.H.; Soltanian-Zadeh, H.: Automatic recognition of five types of white blood cells in peripheral blood. Comput. Med. Imaging Graph. 35(4), 333–43 (2011)
Huang, D.C.; Hung, K.D.; Chan, Y.K.: A computer assisted method for leukocyte nucleus segmentation and recognition in blood smear images. J. Syst. Softw. 85(9), 2104–18 (2012)
Ramesh, N.; Dangott, B.; Salama, M.E.; Tasdizen, T.: Isolation and two-step classification of normal white blood cells in peripheral blood smears. J. Pathol. Inform. 3(1), 13 (2012)
Na, L.; Chris, A.; Mulyawan, B.: A combination of feature selection and co-occurrence matrix methods for leukocyte recognition system. J. Softw. Eng. Appl. 5(12), 101 (2013)
Rezatofighi, S.H.; Soltanian-Zadeh, H.: Automatic recognition of five types of white blood cells in peripheral blood. Comput. Med. Imaging Graph. 35(4), 333–343 (2011)
Xie, E.; McGinnity, T.M.; Wu, Q.: Automatic extraction of shape features for classification of leukocytes. In: 2010 International Conference on Artificial Intelligence and Computational Intelligence (AICI), 2010 Oct 23, vol. 2, pp. 220–224. IEEE (2010)
Ghosh, M.; Das, D.; Mandal, S.; Chakraborty, C.; Pala, M., Maity, A.K.; Pal, S.K.; Ray, A.K.: Statistical pattern analysis of white blood cell nuclei morphometry. In: Students’ Technology Symposium (TechSym), 2010 IEEE, pp. 59–66. IEEE (2010)
Rodrigues, P.; Ferreira, M.; Monteiro, J.: Segmentation and classification of leukocytes using neural networks: a generalization direction. In: Prasad, B., Prasanna, S.R.M. (eds.) Speech, Audio, Image and Biomedical Signal Processing Using Neural Networks, vol. 83, pp. 373–396. Springer, Berlin (2008)
Yampri, P.; Pintavirooj, C.; Daochai, S.; Teartulakarn, S.: White blood cell classification based on the combination of eigen cell and parametric feature detection. In: 2006 1ST IEEE Conference on Industrial Electronics and Applications, 2006 May 24, pp. 1–4. IEEE (2006)
Piuri, V.; Scotti, F.: Morphological classification of blood leucocytes by microscope images. In: 2004 IEEE International Conference on Computational Intelligence for Measurement Systems and Applications, 2004. CIMSA. 2004 Jul 14, pp. 103–108. IEEE (2004)
Bikhet, S.F.; Darwish, A.M.; Tolba, H.A.; Shaheen, S.I.: Segmentation and classification of white blood cells. In: 2000 IEEE International Conference on Acoustics, Speech, and Signal Processing, 2000. ICASSP’00. Proceedings, vol. 4, pp. 2259–2261. IEEE (2000)
Bacusmber, J.W.; Gose, E.E.: Leukocyte pattern recognition. IEEE Trans. Syst. Man Cybern. 2(4), 513–26 (1972)
Young, I.T.: The classification of white blood cells. IEEE Trans. Biomed. Eng. 4, 291–8 (1972)
Adjouadi, M.; Zong, N.; Ayala, M.: Multidimensional pattern recognition and classification of white blood cells using support vector machines. Part. Part. Syst. Charact. 22(2), 107–18 (2005)
Sarrafzadeh, O.; Rabbani, H.; Talebi, A.; Banaem, H.U.: Selection of the best features for leukocytes classification in blood smear microscopic images. In: SPIE Medical Imaging 2014 Mar 20, pp. 90410P–90410P. International Society for Optics and Photonics (2014)
Tabrizi, P.R.; Rezatofighi, S.H.; Yazdanpanah, M.J.: Using PCA and LVQ neural network for automatic recognition of five types of white blood cells. In: 2010 Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), 2010 Aug 31, pp. 5593–5596. IEEE (2010)
Stadelmann, J.V.; Spiridonov, I.N.: Automated classification of leukocytes in blood smear images. Biomed. Eng. 1, 1–5 (2012)
Suapang, P.; Chivaprecha, S.: Automatic leukocyte classification. Int. J. Appl. 8(1), 39–46 (2015)
Mirčić, S.; Jorgovanović, N.: Automatic classification of leukocytes. J. Autom. Control 16(1), 29–32 (2006)
Ferri, M.; Lombardini, S.; Pallotti, C.: Leukocyte classifications by size functions. In: Proceedings of the Second IEEE Workshop on Applications of Computer Vision, 1994, pp. 223–229. IEEE (1994)
Hiremath, P.S.; Bannigidad, P.; Geeta, S.: Automated identification and classification of white blood cells (leukocytes) in digital microscopic images. In: IJCA special issue on “recent trends in image processing and pattern recognition” RTIPPR, pp. 59–63 (2010)
Azar, A.T.; El-Said, S.A.; Balas, V.E.; Olariu, T.: Linguistic hedges fuzzy feature selection for differential diagnosis of Erythemato-Squamous diseases. Soft Comput. Appl. 195, 487–500 (2013)
Rawat, J.; Singh, A.; Bhadauria, H.S.; Virmani, J.; Devgun, J.S.: Computer assisted classification framework for prediction of acute lymphoblastic and acute myeloblastic leukemia. Biocybern. Biomed. Eng. 37, 637–654 (2017)
Mohamed, M.: Image dataset with ground truth images and code. Retrieved on 1-May-2015; MatlabFile exchange from: http://www.mathworks.com/matlabcentral/fileexchange/36634-an-efficient-technique-for-white-blood-cellsnuclei (2012)
Rawat, J.; Bhadauria, H.S.; Singh, A.; Virmani, J.: Review of leukocyte classification techniques for microscopic blood images. In: 2015 2nd International Conference on Computing for Sustainable Global Development (INDIACom), pp. 1948–1954. IEEE (2015)
Putzu, L.; Di Ruberto, C.: White blood cells identification and classification from leukemic blood image. In: Proceedings of the IWBBIO International Work-Conference on Bioinformatics and Biomedical Engineering, pp. 99–106 (2013)
Theerapattanakul, J.; Plodpai, J.; Pintavirooj, C.: An efficient method for segmentation step of automated white blood cell classifications. In: 2004 IEEE Region 10 Conference TENCON 2004, pp. 191–194. IEEE (2004)
Otsu, N.: A threshold selection method from gray-level histograms. Automatica 11(285–296), 23–7 (1975)
Gonzalez, R.C.; Woods, R.E.; Eddins, S.L.: Digital image processing using MATLAB, 3rd edn. Prentice Hall, Upper Saddle River, New Jersey (2004)
Rawat, J.; Singh, A.; Bhadauria, H.S.; Kumar, I.: Comparative analysis of segmentation algorithms for leukocyte extraction in the acute Lymphoblastic Leukemia images. In: 2014 International Conference on Parallel, Distributed and Grid Computing (PDGC), pp. 245–250. IEEE (2014)
Rawat, J.; Singh, A.; Bhadauria, H.S.: An approach for leukocytes nuclei segmentation based on image fusion. In: 2014 IEEE International Symposium on Signal Processing and Information Technology (ISSPIT), pp. 000456–000461. IEEE (2014)
Agaian, S.; Madhukar, M.; Chronopoulos, A.T.: Automated screening system for acute myelogenous leukemia detection in blood microscopic images. IEEE Syst. J. 8(3), 995–1004 (2014)
Madhukar, M.; Agaian, S.; Chronopoulos, A.T.: Deterministic model for acute myelogenous leukemia classification. In: 2012 IEEE International Conference on Systems, Man, and Cybernetics (SMC), pp. 433–438. IEEE (2012)
Baker, Q.B.; Balhaf, K.: Exploiting GPUs to accelerate white blood cells segmentation in microscopic blood images. In: 2017 8th International Conference on Information and Communication Systems (ICICS), pp. 136–140 (2017)
Chan, T.F.; Vese, L.A.: Active contours without edges. IEEE Trans. Image Process. 10(2), 266–77 (2001)
Yang, M.; Kpalma, K.; Ronsin, J.: A survey of shape feature extraction techniques. In: Pattern Recognition, pp. 43-90. InTech (2008)
Haralick, R.M.; Shanmugam, K.; Dinstein, I.: Textural features for image classification. IEEE Trans. Syst. Man Cybern. 3(6), 610–621 (1973)
Rawat, J.; Singh, A.; Bhadauria, H.S.; Virmani, J.; Devgun, J.S.: Classification of acute lymphoblastic leukaemia using hybrid hierarchical classifiers. Multimedia Tools Appl. 76(18), 19057–19085 (2017)
Rawat, J.; Singh, A.; Bhadauria, H.S.; Virmani, J.: Computer aided diagnostic system for detection of leukemia using microscopic images. Procedia Comput. Sci. 1(70), 748–56 (2015)
Abenius, T.: Classification of cell images using MPEG-7-influenced descriptors and support vector machines in cell morphology. arXiv preprint arXiv:0812.2309. (2008 Dec 12).
Kriti,; Virmani, J.: Breast density classification using Laws’ mask texture features. Int. J. Biomed. Eng. Technol. 19(3), 279–302 (2015)
Laws, K.I.: Rapid texture identification. In: 24th Annual Technical Symposium, pp. 376–381. International Society for Optics and Photonics (1980 Dec 23)
Lee, C.C.; Chen, S.H.: Gabor wavelets and SVM classifier for liver diseases classification from CT images. In: SMC’06. IEEE International Conference on Systems, Man and Cybernetics, 2006, vol. 1, pp. 548–552. IEEE (2006 Oct 8 )
Han, Z.Y.; Gu, D.H.; Wu, Q.E.: Feature extraction for color images. In: Hussain, A. (ed.) Electronics, Communications and Networks V, vol. 382, pp. 215–221. Springer, Singapore (2016)
Cetisli, B.: Development of an adaptive neuro-fuzzy classifier using linguistic hedges: part 1. Expert Syst. Appl. 37(8), 6093–101 (2010)
Cetisli, B.: The effect of linguistic hedges on feature selection: part 2. Expert Syst. Appl. 37(8), 6102–8 (2010)
Kher, R.; Pawar, T.; Thakar, V.; Shah, H.: Physical activities recognition from ambulatory ECG signals using neuro-fuzzy classifiers and support vector machines. J. Med. Eng. Technol. 39(2), 138–52 (2015)
Do, Q.H.; Chen, J.F.: A neuro-fuzzy approach in the classification of students’ academic performance. Comput. Intell. Neurosci. 1(2013), 6 (2013)
Jang, J.S.: ANFIS: adaptive-network-based fuzzy inference system. IEEE Trans. Syst. Man Cybern. 23(3), 665–85 (1993)
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Rawat, J., Singh, A., Bhadauria, H.S. et al. Leukocyte Classification using Adaptive Neuro-Fuzzy Inference System in Microscopic Blood Images. Arab J Sci Eng 43, 7041–7058 (2018). https://doi.org/10.1007/s13369-017-2959-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13369-017-2959-3