Abstract
Four new phenolic amides, 4-O-methylgrossamide (1), (E)-2-(4,5-dihydroxy-2-{3-[(4-hydroxyphenethyl)amino]-3-oxopropyl}-phenyl)-3-(4-hydroxy-3-methoxyphenyl)-N-(4-hydroxyphenethyl)acryl-amide (2), (Z)-lyciumamide C (3), (Z)-thoreliamide B (4), together with thirteen known phenolic amides were identified from the stem of Lycium barbarum. The structures of the new compounds were determined by spectroscopic methods. All compounds were evaluated for their anti-cancer activities against human glioma stem cell lines.
Graphical Abstract

Similar content being viewed by others
1 Introduction
Lycium barbarum had a long history of usage as a traditional herbal medicine and functional food in Asian countries [1]. Besides, its fruits known as goji or wolf berries, were beneficial to human health and very important agricultural products [2, 3]. Modern pharmacological studies indicated L. barbarum possessed widely health-promoting and medical effects, including antioxidant [4], lipotropic [5], hepatic function protecting effects [5], immunomodulatory properties [6], antiaging [7], anticancer activities [8,9,10,11] and so on. Phytochemical studies showed that phenolic amides were not only characteristic compounds but also abundant ones in L. Barbarum [2]. Phenolic amides, originating from the condensation of cinnamic acid derivatives and tyramines, octopamines or aliphatic amines [2, 12], had been reported a range of biological activities, like antioxidant [4], antiobesity [13], cytotoxicity [14], anti-inflammatory activity [15] and potent inhibitors of de novo nucleotide biosynthesis [16], and they also seemed to play an important role in plant defense against pathogens [12]. Continuation of our study on the phenolic amides had led to the isolation of four new phenolic amides, 4-O-methylgrossamide (1), (E)-2-(4,5-dihydroxy-2-{3-[(4-hydroxyphenethyl)amino]-3-oxopropyl}-phenyl)-3-(4-hydroxy-3-methoxyphenyl)-N-(4-hydroxyphenethyl)acryl-amide (2), (Z)-lyciumamide C (3), (Z)-thoreliamide B (4), together with thirteen known phenolic amides (Fig. 1) from L. barbarum. The known compounds were identified as grossamide (5) [17], lyciumamide C (6) [4], (Z)-3-{(2,3-trans)-2-(4-hydroxy-3-methoxy-phenyl)-3-hydroxymethyl-2,3-dihydrobenzo[1, 4]-dioxin-6-yl}-N-(4-hydroxyphenethyl)acrylamide (7), [18] (E)-3-{(2,3-trans)-2-(4-hydroxy-3-methoxyphenyl)-3-hydroxy-methyl-2,3-dihydrobenzo[1, 4]dioxin-6-yl}-N-(4-hydroxyphenethyl)acryl-amide (8) [18], (E)-thoreliaide B (9) [19], cannabisin E (10) [20], cannabisin D (11) [21], 1,2-dihydro-6,8-dimethoxy-7-hydroxyl-(3,5-dimethoxy-4-hydroxyphenyl)-N′,N-2-bis[2-(4-hydroxyphenyl)ethyl]-2,3-naphthal-enedicarboxamide (12) [22], cannabisin G (13) [20], N-E-p-coumaroyl tyramine (14) [23], N-E-caffeoyl tyramine (15) [24], N-E-feruloyl tyramine (16) [25], N-E-feruloyl octopamine (17) [25], by comparison with the data in the literature values. All of the compounds were evaluated for anti-cancer activity against human glioma stem cell lines, and compounds 1 and 5 exhibited moderate anti-cancer activities. Herein the isolation, structural elucidation and the bioactivity of the phenolic compounds were reported.
2 Results and Discussion
Compound 1 was obtained as a white powder. Its molecular formula was determined to be C37H38N2O8 on the basis of its 13C NMR and HRESIMS (m/z 639.2721 [M+H]+, calcd. for C37H39N2O8, 639.2701), suggesting 20 degrees of unsaturation. The UV maxima (249, 289 and 320 nm) showed the existence of aromatic rings [19]. The IR spectrum indicated the presence of OH (3387 cm−1), aromatic rings (1651, 1605, 1517 cm−1), and C–N bond (1265 cm−1) groups [14]. In the 1H NMR spectrum, four pairs symmetrical aromatic protons signals at δ H 7.04 (2H, d, J = 8.4 Hz), 6.71 (2H, d, J = 8.4 Hz), 7.01 (2H, d, J = 8.5 Hz), and 6.69 (2H, J = 8.5 Hz) were readily assigned to two AA′BB′ system of 1 [26]. Four pairs of vicinal methylenes protons signals at δ H 2.67 (4H, t, J = 5.1 Hz), 3.36 (4H, t, J = 5.1 Hz), along with four methylenes carbons signals at δ C 34.4 (t), 40.9 (t), 34.2 (t), and 40.8 (t) were the characteristic resonances for two –NHCH2CH2– groups of 1 [19]. In the 13C NMR spectrum, two carbonyl carbon resonances at δ C 169.4 (s) and 165.2 (s) were readily assigned to two amidic moieties, which indicated by HMBC correlations from δ H 5.93 (1H, d, J = 8.1 Hz, H-7) and 3.36 (2H, t, J = 5.1 Hz, H-8″) to δ C 169.4 (s, C-9), from δ H7.38 (1H, d, J = 15.7 Hz, H-7′) and 3.36 (2H, t, J = 5.1 Hz, H-8‴) to δ C 165.2 (s, C-9′). According to these 1D NMR data (Table 1), compound 1 was readily identified as a phenolic amide with two tyramine moieties [18]. Detailed analysis of 1D NMR spectra of 1 displayed similarities to those of 5 [27], except for a methoxy at C-4 in 1 instead of a hydrogen at C-4 in 5, which indicated by HMBC correlations from δ H 3.76 (3H, s, 4-OCH 3), 6.85 (1H, dd, J = 8.4, 2.0 Hz, H-6) and 6.94 (1H, d, J = 2.0 Hz, H-2) to δ C 149.0 (s, C-4), from δ H 3.78 (3H, s, 3-OCH 3) and 6.98 (1H, d, J = 8.4 Hz, H-5) to δ C 148.9 (s, C-3).
The large coupling constants of H-7′ with H-8′ (J = 15.7 Hz) suggested the configuration of the double bond at C-7′/C-8′ was E [17]. The configuration of H-7 with H-8 was threo, which was assigned by the ROESY correlations of H-8 and H-6 (Fig. 3), while there was no REOSY correlation between H-7 and H-8 [28]. The absolute configuration at C-7 and C-8 were 7R, 8S, respectively, which determined by the negative cotton effect at 257 nm (Δε − 0.45) observed in circular dichroic spectrum [28,29,30,31]. Detailed analysis of 2D NMR data (HSQC, HMBC, ROESY) established the structure of 1 to be as shown, named 4-O-methylgrossamide.
Compound 2 was obtained as a yellow powder. Its molecular formula C35H36N2O8 was established by 13C NMR and positive HR–ESI–MS data ([M+Na]+ 635.2368, calcd. for C35H36N2O8Na, 635.2364). The IR spectrum absorptions showed the existence of OH (3422 cm−1), conjugated C=O (1636 cm−1) and Ph (1614 and 1514 cm−1) groups [19]. In the 1D-NMR spectrum, four pairs symmetrical aromatic protons at δ H 6.94 (2H, d, J = 8.4 Hz), 6.68 (2H, d, J = 8.4 Hz), 6.91 (2H, d, J = 8.4 Hz), and 6.65 (2H, d, J = 8.4 Hz) suggested the presence of two AA′BB′ systems [26]. Four pairs of vicinal methylenes protons signals at δ H 2.66 (2H, t, J = 7.5 Hz), 3.41 (2H, t, J = 7.5 Hz), 2.58 (2H, t, J = 7.4 Hz), and 3.20 (2H, t, J = 7.4 Hz), along with two carbonyl carbons at δ C 171.4 (s), and 166.8 (s) were the characteristic resonances for two –CONHCH2CH2– moieties, which were supported by HMBC correlations of δ H 7.57 (1H, s, H-7), 3.41 (2H, t, J = 7.5 Hz, H-8″) with δ C 166.8 (s, C-9), of δ H 2.43 (1H, m, H-7′), 3.20 (2H, t, J = 7.4 Hz, H-8‴) with δ C 171.4 (s, C-9′) [19]. Analysis of the 1H, 13C NMR data (Table 1) indicated that 2 was similar to those of (E)-2-(4,5-dihydroxy-2-{3-[(4-hydroxyphenethyl)amino]-3-oxopropyl}-phenyl)-3-(4-hydroxy-3,5-dimethyoxyphenyl)-N-(4-hydroxyphenethyl)-acrylamide [18], except for the missing of –OCH3 at C-5 in 2, as supported by the highfield shifted at C-5 (from δ C 148.7 in the forementioned to δ C 115.2 in 2) and the HMBC correlations from 6.66 (1H, d, J = 8.3 Hz, H-5) with δ C 126.5 (s, C-1) and 147.4 (s, C-3). The only methoxyl was substituted at C-3, which was supported by the HMBC correlations of δ H 3.37 (3H, s), 6.66 (1H, d, J = 8.3 Hz, H-5) with δ C 147.4 (s, C-3). The HMBC correlations of δ H 2.43 (1H, m, H-7′) with δ C 171.4 (s, C-9′), 131.4 (s, C-6′), and 116.7 (d, C-2′), of δ H 2.13 (2H, td, J = 9.4, 6.5 Hz, H-8′) with δ C 130.7 (s, C-1′), suggested the –CH2CH2– fragment was connected between C-1′ and C-9′ (Fig. 2). ROESY correlation of H-6 with H-5′ (Fig. 3) suggested two benzene rings, connected to C-7 and C-8 respectively, were on the same side, further confirmed the double bond of C-7/C-8 was E [18]. Hence, Detailed analysis of 2D NMR data established the structure of compound 2 to be as shown, named (E)-2-(4,5-dihydroxy-2-{3-[(4-hydroxyphenethyl)amino]-3-oxopropyl}phenyl)-3-(4-hydroxy-3-methoxy-phenyl)-N-(4-hydroxyphenethyl)acrylamide.
The other phenolic amide, 3, obtained as a white powder, with the same R f value on silica plate as that of 6, showed identical physical data in the HRESIMS and IR spectra, indicating the existence of the same molecular formula and functional groups as in 6 [4]. The 1D-NMR displayed an AA′BB′ system [26] [δ H 6.96 (2H, d, J = 8.4 Hz), and 6.68 (2H, d, J = 8.5 Hz), along with δ C 130.8 (d), and 116.4 (d)], five proton signals [(δ H 7.22 (1H, d, J = 1.7 Hz), 7.05 (1H, s), 6.93 (1H, d, J = 2.0 Hz), 6.81 (1H, dd, J = 8.1, 2.0 Hz) and 6.75 (1H, d, J = 8.1 Hz)] suggested the existence of two aromatic rings [18]. The 13C NMR also showed an amidic carbon [δ C 170.3 (s, C-9) and δ C 42.5 (t, C-8″)], which indicated by HMBC correlations from δ H 6.66 (1H, d, J = 12.6 Hz, H-7) and 3.31 (2H, t, J = 7.5 Hz, H-8″) to δ C 170.3 (s, C-9) (Fig. 2).
Detailed analysis of 1D, 2D NMR spectral data (Table 2) suggested that the planar structure of 3 was the same as 6. The visible difference was that the configuration of the double bond at C-7/C-8 in 3 was Z, which was suggested by coupling constants (J = 12.6 Hz) between H-7 and H-8 [4, 25]. The NOESY correlations of H-7′ and H-8′ suggested the configuration of H-7′/H-8′ was erythro (Fig. 3) [4], which also supported by the specific optical rotation of 3 \([[\upalpha]_{\text{D}}^{25}\) − 5.0° (c 0.11, MeOH)] was the same side as that of 6 \([[\upalpha]_{\text{D}}^{22}\) − 7.9° (c 0.34, MeOH)] [4]. Other parts of the structure were identical to those of 6 by detailed analysis of its 2D NMR spectra. Thus, the structure of 3 was established and named Z-lyciumamide C.
The aromatic rings of the phenylpropanoid units exhibited various oxygenation patterns, and it had been summarized that the deoxygenation patterns might involve the 3,4-, 2,4-, 2,5- and 2,6-positions [4, 17, 19,20,21], but seldom the 3,5-positions [18]. Some compounds which had 3,5-positions substituents always been proven to be 3,4- 2,4- or 2,5-positions substituents, owing to the existence of “deceptively simple” protons signals when changed the deuterated solvent or temperature [32]. Interestingly, in our experiment, the “deceptively simple” protons signals that exhibited two broad singlets with approximate integrations of 1:2 for H-2′ and for H-5′, H-6′ were found when using DMSO-d 6, and they would be a set of proton signals with a m-coupling constant for H-2′ (d, J = 2.0 Hz), an o-coupling constant for H-5′ (d, J = 8.1 Hz), and o-,m-coupling constants for H-6′ (dd, J = 8.1, 2.0 Hz) in 3, and with a m-coupling constant for H-2′ (d, J = 2.0 Hz), an o-coupling constant for H-5′ (d, J = 8.2 Hz), and o-,m-coupling constants for H-6′ (dd, J = 8.2, 2.0 Hz) in 6 when using CD3OD (SI, Figs. S37, S38).
Compound 4 was obtained as faint yellow powder. Its molecular formula C28H29NO8 was established by 13C NMR and positive ESIMS m/z 508 [M+H]+. The UV maxima (207, 240 and 278 nm) showed the existence of aromatic rings and IR bands (3421, 1648, 1614, 1512, and 1274 cm−1) displayed aromatic rings, hydroxyl functions and a C–N bond [18]. Analysis of the 1H, 13C NMR data (Table 1) revealed that compounds 4 and 9 are structurally similar except for the configuration of the double bone at C-7/C-8. The coupling constants (J = 12.6 Hz) between H-7 and H-8 suggested the configuration of the double bone at C-7/C-8 in 4 was Z [21]. The 1H–1H COSY correlations of δ H 4.88 (1H, d, J = 8.1 Hz, H-7′)/4.07 (1H, ddd, J = 8.1, 4.3, 2.5 Hz, H-8′)/3.71 (1H, dd, J = 12.5, 2.5 Hz, H-9′)/3.48 (1H, dd, J = 12.5, 4.3 Hz, H-9′) revealed the presence of –CH(7′)–CH(8′)–CH2(9′)– fragment, also supported by HMBC correlations of δ H 4.88 (1H, d, J = 8.1 Hz, H-7′), 4.07 (1H, ddd, J = 8.1, 4.3, 2.5 Hz, H-8′) to δ C 106.0 (d, C-2′, 6′) (Fig. 2). The relative configuration of H-7′/H-8′ was threo orientation, for the large coupling constant (J = 8.1 Hz) between H-7′ and H-8′, along with ROESY correlation of H-2′ and H-6′ with H-8′ and there was no NOE correlation between H-7′ and H-8′ [33,34,35] (Fig. 3). Other parts of the structure were identical to those of 9 by detailed analysis of its 2D NMR spectra. Thus, compound 4 was assigned as (Z)-thoreliamide B.
A literature survey shows that the E-isomers of this type of compounds are widespread in some genera and small amount of Z-isomers [12, 18]. In this paper, three pairs of Z-E-isomers (3 and 6; 4 and 9; 7 and 8) were reported. Due to their different retention times on the Rp-C18 column, every pair of isomers was separated by HPLC.
All the compounds were evaluated for their bioactivity against two human glioma stem cell lines (GSC-3# and GSC-12#), by the cell viability assay and phenotypic screening. The results showed that compound 5 exhibited the moderate cytotoxicity against GSC-3# and GSC-12# at the concentration of 10 μg/mL (Fig. 4a), and the IC50 values were 6.40 and 5.85 μg/mL respectively (Fig. 5). Compound 1 showed the moderate cytotoxicity against GSC-3# and GSC-12# at the concentration of 25 μg/mL (Fig. 4b), and the IC50 values were 28.51 and 19.67 μg/mL respectively (Fig. 5).
3 Experimental Section
3.1 General Experimental Procedures
Optical rotations were measured on a JASCO P-1020 polarimeter. UV spectra were detected on a SHMADZU UV-2401PC spectrometer. IR spectra were determined on a Bruker FT-IR Tensor-27 infrared spectrophotometer with KBr disks. 1D and 2D NMR spectra were recorded on Bruker DRX-400, DRX-500, and DRX-600 spectrometers using TMS as an internal standard. Chemical shifts (δ) were expressed in ppm with reference to the solvent signals. ESI–MS and EI–MS (HR–EI–MS) analysis were carried out on Waters Xevo TQS and Waters AutoSpec Premier P776 mass spectrometers, respectively. Semi-preparative HPLC was performed on a Waters 600 with a COSMOSIL C18 (10 × 250 mm) column. Silica gel (100–200 and 200–300 mesh, Qingdao Marine Chemical Co., Ltd., People’s Republic of China), and MCI gel (75–150 μm, Mitsubishi Chemical Corporation, Tokyo, Japan) were used for column chromatography. Fractions were monitored by thin-layer chromatography (TLC) (GF254, Qingdao Marine Chemical Co., Ltd.), and spots were visualized by 10% sulfuric acid ethanol solution and dragendorff reagent.
3.2 Plant Material
The stem of L. barbarum were collected from Zhongning county of Ningxia province, where the Chinese wolfberries producing, and identified by Mr. Jianfei Liu, Lanzhou Institute of Chemical Physics, CAS. A voucher specimen (No. 20150602) has been deposited in the Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, China.
3.3 Extraction and Isolation
The air-dried and powdered sample (10.0 kg) was extracted with 85% aqueous EtOH (40 L × 3) under reflux conditions for 3 h each time, and the solvent was evaporated in vacuum. The residue (607 g) was suspended in H2O and extracted with EtOAc (each 2 L × 3). The EtOAc layer (224 g) was passed over a silica gel column, eluting with CHCl3–Me2CO (1:0 to 0:1) to give nine fractions. Fraction VI (22 g) was chromatographed on silica gel, eluted with CHCl3–MeOH (20:1 to 6:1) to give 12 (568.9 mg) and the residue, then the latter was further purified over HPLC to afford 2 (4.86 mg, t R 18 min; CH3CN/H2O 20:80, 3 mL/min), and 11 (15.5 mg, t R 23.5 min; CH3CN/H2O 20:80, 3 mL/min). Fraction VII (62 g) was separated on a MCI column eluted successively with MeOH/H2O (10:1–5:1) to afford seven subfractions VII-1–7 and one white needle crystal 16 (7283.1 mg, in chloroform). Subfraction VII-2 was separated by Sephadex LH-20 column, eluted with MeOH, to afford compound 14 (1154.4 mg). Subfraction VII-3 was separated by RP-MPLC, eluted with MeOH/H2O (20:100–55:100) to get subfractions VII-3-1 and VII-3-2. Subfraction VII-3-1 was chromatographed on silica gel, eluted with CHCl3–MeOH (20:1–5:1) to give 15 (535.7 mg), 17 (88.0 mg) and 13 (168.3 mg). Subfraction VII-3-2 was purified over HPLC to afford 10 (23.1 mg, t R 28 min; CH3CN/H2O 28:72, 3 mL/min). Subfraction VII-4 was separated on the column of polyamide, eluted with MeOH/H2O (45:100–60:100), to yield compound 5 (185.0 mg) and a mixture. Further separation of the mixture by on a silica gel column (CHCl3/MeOH, 20:1–9:1) yielded compound 1 (43.5 mg). Subraction VII-5 firstly separated by Sephadex LH-20 column to obtain two major subfraction VII-5-1 and VII-5-2, and the former was purified by HPLC to afford 4 (4.2 mg, t R 16 min; CH3CN/H2O 35:65, 3 mL/min) and 9 (5.6 mg, t R 21.5 min; CH3CN/H2O 35:65, 3 mL/min). Separation of subfraction VII-5-2 by silica gel column (CHCl3/MeOH, 20:1–9:1) to obtain subfractions VII-5-2a and VII-5-2b. Purified of subfraction VII-5-2a by HPLC afford 6 (54.3 mg, t R 18 min; CH3CN/H2O 38:62, 3 mL/min), 3 (43.2 mg, t R 23 min; CH3CN/H2O 38:62, 3 mL/min). Subfraction VII-5-2b was separated by Sephadex LH-20 column (elution with MeOH), and then chromatographed on silica gel column (CHCl3/MeOH, 10:1), further purified by HPLC to afford 7 (12.5 mg, t R 20 min; CH3CN/H2O 38:62, 3 mL/min) and 8 (7.8 mg, t R 25 min; CH3CN/H2O 38:62, 3 mL/min).
3.3.1 4-O-methylgrossamide (1)
White powder; \([\upalpha]_{\text{D}}^{25}\) − 16.8° (c 0.11, DMSO); UV (MeOH) λ max (log ε) 249 (4.2), 289 (4.4), 320(4.4); IR (KBr) ν max 3387, 1651, 1605, 1517, 1265 cm−1; 1H NMR (DMSO- d 6, 600 MHz) and 13C NMR (DMSO- d 6, 150 MHz) data, see Table 1; positive ESIMS m/z 639 [M+H]+; positive HRESIMS m/z 639.2721 [M+H]+ (calcd. for C37H39N2O8, 639.2701).
3.3.2 (E)-2-(4,5-dihydroxy-2-{3-[(4-hydroxyphenethyl)amino]-3-oxopro-pyl}phenyl)-3-(4-hydroxy-3-methoxyphenyl)-N-(4-hydroxyphenethyl)-acrylamide (2)
Yellow powder; \([\upalpha]_{\text{D}}^{19}\) − 2.3° (c 0.47, MeOH); UV (MeOH) λ max(log ε) 203 (4.7), 217 (4.6), 289 (4.1), 324 (4.1); IR (KBr) ν max 3422, 2927, 1636, 1614, 1514, 1449, 1262 cm−1; 1H NMR (DMSO- d 6, 600 MHz) and 13C NMR (DMSO- d 6, 150 MHz) data, see Table 1; positive HRESIMS [M+Na]+ 635.2368 (calcd. for C35H36N2O8Na, 635.2364).
3.3.3 Z-lyciumamide C (3)
White powder; \([\upalpha]_{\text{D}}^{26}\) − 5.0° (c 0.11, MeOH); UV (MeOH) λ max (log ε) 203 (4.6), 225 (4.5), 286 (4.2), 304 (4.2); IR (KBr) ν max 3408, 1647, 1608, 1516, 1273, 1216 cm−1; 1H NMR (CD3OD, 500 MHz) and 13C NMR (CD3OD, 125 MHz) data, see Table 2; positive ESIMS m/z 492 [M+H]+; positive HRESIMS m/z 492.2027 [M+H]+ (calcd. for C28H30NO7, 492.2017).
3.3.4 Z-thoreliamide B (4)
Faint yellow powder; \([\upalpha]_{\text{D}}^{21}\) − 11.1° (c 0.11, MeOH); UV (MeOH) λ max (log ε) 207 (4.7), 240 (4.3), 278 (4.0); IR (KBr) ν max 3421, 1648, 1614, 1512, 1274, 1219 cm−1; 1H NMR (CD3OD, 600 MHz) and 13C NMR (CD3OD, 150 MHz) data, see Table 1; positive ESIMS m/z 508 [M+H]+; negtive HRESIMS m/z 506.1825 [M−H]− (calcd. for C28H28NO8, 506.1820).
3.4 Anticancer Activities
GSC-3# and GSC-12# were human glioma stem cell lines that were established by Kunming institute of zoology from two human glioblastoma multiform samples. The glioma stem cell was cultured in serum-free medium DMEM F12 supplied with 1xB27 and 50 ng/mL EGF, BFGF and 1% penicillin/streptomycin. The glioma stem cells were seeded in the laminin pre-coating dishes and cultured in 37 °C, 5% CO2 incubator. Cell viability assay was performed by the MTS method as previously described. GSCs were digested and counted, seeded in laminin pre-coating 96-well-plate with 20000 cells/well. The compounds were added with a serial gradient concentration (40, 20, 10, 5, 2.5, 1.25, 0.625, 0.3125 μg/mL) and cultured in cell incubator for 72 h. MTS reagent was diluted 1:5 with fresh medium and mixed well. The old medium was removed and subsequently the fresh medium was added with 100 μL/well. The cells were incubated for 1.5 h. Absorbance was measured by Hybrid Reader (BioTek synergy H1) at 490 nm. The cell viability was evaluated by percentage compared with DMSO control group. The half-maximal inhibitory concentration (IC50) was measured and calculated by Graph Pad Prism 5 software.
4 Supporting Information
1D, 2D NMR spectra, ESIMS/MS, HRESIMS, UV and IR of compounds 1–4 and influence of deuterated solvent on the 1H NMR spectra of compounds 3 and 6 are available).
References
O. Potterat, Planta Med. 76, 7–19 (2010)
J. Zhang, S. Guan, J. Sun, T. Liu, P. Chen, R. Feng, X. Chen, W. Wu, M. Yang, D.A. Guo, Anal. Bioanal. Chem. 407, 581–595 (2015)
M. Forino, L. Tartaglione, C. Dell’Aversano, P. Ciminiello, Food Chem. 194, 1254–1259 (2016)
K. Gao, D.W. Ma, Y. Cheng, X.R. Tian, Y.Y. Lu, X.Y. Du, H.F. Tang, J.Z. Chen, J. Agric. Food Chem. 63, 1067–1075 (2015)
Y. Kajimoto, S. Kurokawa, Folia Pharmacol. 56, 151 (1960)
L.W. Xie, S.X. Li, Y.X. Xie, Y. Pan, R. Yu, X.H. Cheng, China J. Chin. Mater. Med. 39, 689–694 (2014)
R.C. Chang, K.F. So, Mol. Neurobiol. 28, 643–652 (2008)
F. Mao, B. Xiao, Z. Jiang, J. Zhao, X. Huang, J. Guo, Med. Oncol. 28, 121–126 (2011)
A. Wawruszak, A. Czerwonka, K. Okla, W. Rzeski, Nat. Prod. Res. 30, 1993–1996 (2016)
W.D. Liu, F.X. Yi, Q.S. Hui, Acad. J. China. PLA 34, 179–180 (2013)
N.P. Seeram, L.S. Adams, Y. Zhang, D. Sand, D. Heber, J. Agric. Food Chem. 54, 9329–9339 (2006)
U. Muhlenbeck, A. Kortenbusch, W. Barz, Phytochemistry 42, 1573–1579 (1996)
J. Joven, E. Espinel, A. Rull, G. Aragones, E. Rodriguez-Gallego, J. Camps, V. Micol, M. Herranz-Lopez, J.A. Menendez, I. Borras, Biochim. Biophys. Acta 1820, 894–899 (2012)
K.H. Kim, S.U. Choi, M.W. Son, K.R. Lee, Chem. Pharm. Bull. 58, 1532–1535 (2010)
S. Wang, J.H. Suh, X. Zheng, Y. Wang, C.T. Ho, J. Agric. Food Chem. 65, 364–372 (2017)
T. Pisithkul, T.B. Jacobson, T.J. O’Brien, D.M. Stevenson, D. Amador-Noguez, Appl. Environ. Microbiol. 81, 5761–5772 (2015)
A.M. Seca, A.M. Silva, A.J. Silvestre, J.A. Cavaleiro, F.M. Domingues, C. Pascoal-Neto, Phytochemistry 58, 1219–1223 (2001)
J.X. Zhang, S.H. Guan, R.H. Feng, Y. Wang, Z.Y. Wu, Y.B. Zhang, X.H. Chen, K.S. Bi, D.A. Guo, J. Nat. Prod. 76, 51–58 (2013)
F. Ge, C.P. Tang, Y. Ye, Helv. Chim. Acta 91, 1023–1030 (2008)
I. Sakakibara, Y. Ikeya, K. Hayashi, M. Okada, M. Maruno, Phytochemistry 38, 1003–1007 (1995)
I. Sakakibara, Y. Ikeya, K. Hayashi, H. Mitsuhashi, Phytochemistry 31, 3219–3223 (1992)
M.H. Chaves, N.F. Roque, Phytochemistry 46, 879–881 (1997)
Y. Zhao, Q.S. Zhao, Z.W. Lin, H.D. Sun, H.M. Wu, J.F. Xu, Acta Bot. Sin. 40, 1035–1039 (1998)
D.G. Lee, Y. Park, M.R. Kim, H.J. Jung, Y.B. Seu, K.S. Hahm, E.R. Woo, Biotechnol. Lett. 26, 1125–1130 (2004)
R.R. King, L.A. Calhoun, Phytochemistry 66, 2468–2473 (2005)
X.L. Yan, J.J. Tang, C.S. Passos, A. Nurisso, C.A. Simoes-Pires, M. Ji, H.X. Lou, P.H. Fan, J. Agric. Food Chem. 63, 10611–10619 (2015)
L.P. Santos, M.A.D. Boaventura, D.O. Alaide, Braga, J.M. Cassady. Planta Med. 62, 76 (1996)
J. Sun, Y.F. Gu, X.Q. Su, M.M. Li, H.X. Huo, J. Zhang, K.W. Zeng, Q. Zhang, Y.F. Zhao, J. Li, P.F. Tu, Fitoterapia 98, 110–116 (2014)
M.S.M. Yuen, F. Xue, T.C.W. Mak, H.N.C. Wong, Tetrahedron 54, 12429–12444 (1998)
M. Yoshikawa, T. Morikawa, F.M. Xu, S. Ando, H. Matsuda, Heterocycles 60, 1787–1792 (2003)
L. Xiong, C.G. Zhu, Y.R. Li, Y. Tian, S. Lin, S.P. Yuan, J.F. Hu, Q. Hou, N.H. Chen, Y.C. Yang, J.G. Shi, J. Nat. Prod. 74, 1188–1200 (2011)
Y.N. Yang, H. Zhu, Z. Chen, F. Liu, Y.W. An, Z.M. Feng, J.S. Jiang, P.C. Zhang, J. Nat. Prod. 78, 705–711 (2014)
P.G.M. Wuts, S.S. Bigelow, J. Org. Chem. 47, 2498–2500 (1982)
T. Oshihara, K. Yamaguchi, S. Sakamura, Agric. Biol. Chem. 47, 217–220 (1983)
R. Waibel, G. Benirschke, M. Benirschke, H. Achenbach, Phytochemistry 62, 805–811 (2003)
Acknowledgements
The authors are grateful to agricultural comprehensive development project of science and technology in Ningxia province (Research on Chinese wolfberry active substances and health products), and STS project of Chinese Academy of Sciences for the financial support.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Zhu, PF., Dai, Z., Wang, B. et al. The Anticancer Activities Phenolic Amides from the Stem of Lycium barbarum . Nat. Prod. Bioprospect. 7, 421–431 (2017). https://doi.org/10.1007/s13659-017-0134-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13659-017-0134-x


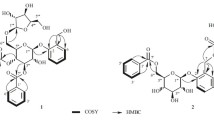



 ) and HMBC (
) and HMBC ( ) correlations for compounds 1–4
) correlations for compounds 1–4

 ) correlations of compounds 1–4
) correlations of compounds 1–4

