Abstract
The novel coronavirus (COVID-19) is continuing its spread across the world, claiming more than 160,000 lives and sickening more than 2,400,000 people as of April 21, 2020. Early research has reported a basic reproduction number (R0) between 2.2 to 3.6, implying that the majority of the population is at risk of infection if no intervention measures were undertaken. The true size of the COVID-19 epidemic remains unknown, as a significant proportion of infected individuals only exhibit mild symptoms or are even asymptomatic. A timely assessment of the evolving epidemic size is crucial for resource allocation and triage decisions. In this article, we modify the back-calculation algorithm to obtain a lower bound estimate of the number of COVID-19 infected persons in China in and outside the Hubei province. We estimate the infection density among infected and show that the drastic control measures enforced throughout China following the lockdown of Wuhan City effectively slowed down the spread of the disease in two weeks. We also investigate the COVID-19 epidemic size in South Korea and find a similar effect of its “test, trace, isolate, and treat” strategy. Our findings are expected to provide guidelines and enlightenment for surveillance and control activities of COVID-19 in other countries around the world.
Similar content being viewed by others
Introduction
In early December 2019, a cluster of pneumonia cases of unknown etiology was reported in Wuhan, a city of 11 million residents in central China. The Chinese Center for Disease Control and Prevention (China CDC) reported a novel coronavirus as the causative agent of this outbreak on January 9, 2020. To contain the spread of the virus, Wuhan, the epicenter of the coronavirus epidemic, has been placed in lockdown since January 23, 2020. The order was later expanded to the entire Hubei province in the next few days, affecting nearly 56 million people. However, it was estimated that 5 million people already left the central Chinese city, as China’s great Lunar New Year migration has already broken across the nation in the first few weeks of January. Some carried with them the new virus that has since spread throughout China and to 215 other countries, claiming almost 750,000 lives and sickening more than 20,000,000 people as of April 10, 2020. After characterizing the outbreak a Public Health Emergency of International Concern in late January, the World Health Organization (WHO) eventually declared COVID-19 as a pandemic on March 11, 2020.
Thus far, early research on the basic reproduction number (R0) of COVID-19 has reported an estimated R0 in the range of 2.2 to 3.6 [1,2,3,4], which means that, on average, each infected person spreads the infection to more than two persons. Therefore, the majority of the population is at risk of infection if no intervention measures were undertaken. The true size of the COVID-19 epidemic remains unknown, as a significant proportion of infected individuals only exhibit mild symptoms or are even asymptomatic.
The intensive care needed to treat COVID-19 patients is adding pressure to the already stressed healthcare system worldwide. A recent report from WHO found that the case fatality rate was 5.8% in Wuhan, compared with 0.7% in the rest of the country during the same time period [5]. The striking difference is mainly due to the sudden surge of severely ill people overwhelming the healthcare system. Hence a timely assessment of the evolving epidemic size is crucial for resource allocation and triage decisions.
In the literature, there are generally three different types of approaches for estimating the number of infected persons. The first approach fits and estimates a model for the incidence rate curve during the observation period and then extrapolates into the future [6, 7]. Although easy to implement, this approach is known to be risky and unreliable due to extrapolating a fitted model outside the range of the observed data. The second approach involves modeling the dynamics of the epidemic of interest. Mathematical models of infectious diseases have a long history [8]. Unfortunately, the deterministic models proposed for general epidemics are complicated, and the stochastic models are even more complicated.
The third method is the back-calculation procedure developed by Brookmeyer and Gail [9, 10], the focus of this work. An obvious advantage of this method is that it requires no assumptions about the number of infected individuals in the population or the proportion of infected individuals or models on the dynamics of the COVID-19 epidemic. We consider applying the back-calculation procedure to estimate the epidemic size. This procedure was originally developed for estimating the number of subjects infected with human immunodeficiency virus (HIV). It projects the observed numbers of confirmed cases to numbers previously infected, where the number of confirmed cases in each time interval follows a multinomial distribution with cell probabilities that can be expressed as a convolution of the distribution of the infection time (termed as infection distribution) and that of the time from infection to diagnosis (termed as incubation distribution). As a result, the problem is reduced to estimating the size of a multinomial population. This approach has been applied to study other infectious diseases such as bovine spongiform encephalopathy epidemic [10, 11].
Direct application of the back-calculation procedure, however, requires the knowledge of the incubation distribution of the disease under study. For HIV, the majority of infected individuals will eventually develop symptoms and get diagnosed; as a result, the incubation time can be reliably estimated. However, the incubation distribution for COVID-19 varies across different regions due to different testing and contacting tracing policies. Hence the naive back-calculation procedure can not be directly applied to COVID-19, especially in the early stage of pandemics. To tackle this problem, we propose a modified back-calculation procedure by imposing a parametric model on the incubation distribution of COVID-19, alleviating the requirement of a known incubation distribution. Similar to its original form, the modified back-calculation procedure can not be used to predict future new cases but it gives a lower bound estimate of the number of confirmed cases in the near future, which is a crucial piece of information to guide decision making on medical resource allocation.
The paper proceeds as follows. In section “Methods”, we introduce our modified back-calculation procedure and the maximum full likelihood estimators for all model parameters. In section “Results”, we apply the proposed procedure to analyze the COVID-19 epidemics outside Hubei province, China (section “COVID-19 epidemics outside Hubei province, China before March 15, 2020”), inside Hubei province (section “COVID-19 epidemics in Hubei province, China”) and in South Korea (section “COVID-19 epidemics in South Korea”). We conclude in section “Conclusion”. Section 4 contains some discussions.
Methods
Let U denote the calendar time of COVID-19 infection for an individual and let T denote the incubation time from infection to diagnosis. Note that cases may be diagnosed through active surveillance testing, not just based on development of symptoms. Therefore an individual is diagnosed before a calendar time \(\tau\) if and only if \(U+T\le \tau\). Suppose the numbers of confirmed cases of COVID-19 in a series of time intervals \([\tau _0, \tau _1)\), \([\tau _1, \tau _2)\), ..., \([\tau _{K-1}, \tau _K)\) are available, where we set \(\tau _0\equiv -\infty\), so that no infection occurred prior to \(\tau _0\), and \(\tau _K\) is the date of the last available report. Assume that the distribution of U (infection distribution) given \(U\le \tau _K\) has a density function \(\phi (u; \varvec{\alpha } )\), \(u\le \tau _K\), where \(\varvec{\alpha }\) is a \(p_\alpha\)-dimensional vector of parameters. Moreover, we assume that, given \(U=u\), T is a nonnegative, continuous random variable with the distribution function \(F_u(t; \varvec{\beta } )\), where \(\varvec{\beta }\) is a \(p_\beta\)-dimensional vector of parameters. To implement the maximum likelihood estimation, we assume that \(F_u(t; \varvec{\beta } )\equiv F(t; \varvec{\beta } )\), that is, the incubation time is independent of the date when the individual was infected. This assumption may not be valid for the confirmed case in Wuhan or other cities in the Hubei province, as the medical resource in the early stage of the epidemic is extremely scarce and thus it may take a long time for infected individuals to be diagnosed. In fact, the surge in the number of confirmed cases in China on February 13 was due to a new diagnosis classification rule for cases in the Hubei province; the RT-PCR test for COVID-19 was not available for many of the previously infected individuals despite having symptoms of pneumonia. On the other hand, this assumption seems reasonable for areas outside of Hubei province as the healthcare system has not been overly stressed.
It follows from the assumption that T independent of U given \(U\le \tau _K\) that the probability of being diagnosed in the time interval \([\tau _{j-1}, \tau _j)\), conditioning on being infected before the last examination time \(\tau _K\), is given by
Let \(\varvec{\theta } =(\alpha ^{\intercal }, \beta ^{\intercal })^{\intercal }\) and define \(\pi _j( \varvec{\theta } ) =\int _{\tau _0}^{\tau _K} \left\{ F(\tau _j-u;\beta ) - F(\tau _{j-1}-u;\beta ) \right\} \phi (u;\alpha )du\).
Depiction of the data structure. U’s are the calendar times of infection and T’s are the incubation times. U’s and T’s are unobserved while the daily numbers of diagnosed cases \(X_1, \ldots , X_K\) in the K time intervals are observed. Here \(\pi _1, \ldots , \pi _K\) are the probabilities of being diagnosed in the K time intervals among those infected before time \(\tau _K\)
Denote by N the (unknown) number of individuals infected before \(\tau _K\) and let \(X_j\), \(j=1,\ldots , K\) be the number of cases diagnosed in the time interval \([\tau _{j-1}, \tau _j)\). Define \(n=\sum _{j=1}^K X_j\), so that a total of \(N-n\) individuals were infected but not diagnosed before \(\tau _K\). Define \(\pi _{K+1}( \varvec{\theta } ) =\hbox {Pr}( U+T \ge \tau _K \mid U\le \tau _K ) =1-\sum _{i=1}^K \pi _{j}( \varvec{\theta } )\). Figure 1 depicts the structure of observed data and model parameters. Write \({\widetilde{ \varvec{X} }}=(X_1, \ldots , X_K, N-n)^{\intercal }\) and \({\widetilde{ \varvec{\pi } }}( \varvec{\theta } )=(\pi _1( \varvec{\theta } ), \ldots , \pi _K( \varvec{\theta } ),\pi _{K+1}( \varvec{\theta } ))^{\intercal }\), so \({\widetilde{ \varvec{X} }}\) follows the multinomial distribution with trial size N and cell-probabilities \({\widetilde{ \varvec{\pi } }}( \varvec{\theta } )\). Given the observed data, \(X_1, \ldots , X_K\), the likelihood function of \((N, \varvec{\theta } )\) is
The corresponding log-likelihood, up to a constant, is
where \(\Gamma (x+1)=x!\) is the Gamma function. We propose to estimate \((N, \varvec{\theta } )\) by the maximizer of likelihood function
Then the expected number of confirmed cases in the time interval \([\tau _{j-1}, \tau _j)\) can be estimated by \({\widehat{N}}\times {\widehat{\pi }}_j({\widehat{ \varvec{\theta } }})\), where
It is worthwhile to point out that the back-calculation algorithm described above estimates the two sets of parameters \(( \varvec{\alpha } , \varvec{\beta } )\) simultaneously; this is different than the original algorithm where the parameters \(\varvec{\beta }\) in the Weibull distribution were replaced by values derived from other studies which provided information about the incubation time [9, 10]. In our analysis, T stands for the duration between infection and laboratory confirmation, whose information plays an important role in guiding medical resource allocation. Since no active surveillance testing and contact tracking were conducted in China before March 15, the length of time to confirmation is expected to be longer than the incubation time, that is, the duration between infection and symptom onset.
We have performed the analysis by incorporating the estimated incubation distribution reported in [12, p. 11], but the model did not fit well. As a result, we decided to estimate the two sets of parameters simultaneously. However, one major consequence of such a strategy is that the distribution of U and T are estimated subject to a location shift factor. To see this, we note that the infection time, U, and the time from infection to diagnosis, T, are not directly observed in the data, and we only get to observe \(U+T\), that is, the time of confirmation. Hence a different set of random variables \(T^*=T-\delta\) and \(U^*=U+\delta\) yield the same distribution as \(T+U\). On the other hand, although the location shift factor \(\delta\) can not be estimated directly from the observed data, we can assess the magnitude of the location shift by comparing the estimated incubation distribution to what reported in the existing literature. Note that, under location shift, the relative difference in time between two landmark time points, such as the peak and lowest point in the infection density or last week compared to this week, can be estimated from the data.
Results
COVID-19 epidemics outside Hubei province, China before March 15, 2020
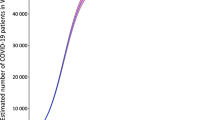
The National Health Commission of the People’s Republic of China has published daily numbers of confirmed cases of COVID-19 since January 20, 2020; see http://www.nhc.gov.cn/xcs/yqtb/list_gzbd.shtml for all news releases. We analyze data from areas outside the Hubei Province during the 8-week period between January 20 and March 15. Note that March 15 was selected because the majority of the newly confirmed cases have been imported cases afterwards. The daily numbers of confirmed cases are graphically depicted in Figure 2A. It can be observed that the daily number of confirmed cases reached its peak on February 15 with 890 new cases, 11 days after the lockdown of Wuhan City on January 23. The spike of 261 new cases on February 20 was due to delayed reports of outbreaks in two prisons.
We modeled the incubation distribution using the Weibull distribution, which has been shown a reasonable approximation for the incubation time in infectious disease research. As argued before, the stationarity assumption for the time to diagnosis, that is, the incubation distribution does not depend on the time of infection, should hold approximately in cases diagnosed outside the Hubei Province. Moreover, for modeling the infection density function \(\phi (t, \varvec{\alpha } )\), we assume a step function with jump discontinuities every 7 days starting from January 20 (day 0), so that the risk of infection is constant within each week. Since the first two cases diagnosed outside of Hubei were reported to have visited Wuhan on January 7 and 9 and developed symptoms on January 13 and 14, respectively, we set \(\tau _0\) to January 1, 2020 to account for the possible infection period. Specifically, the infection density function among cases infected before March 15 is of the form
where \(a_0=-19\), \(a_j=7\times (j-1)\), \(j=1, \ldots , 9\). Note that we require \(\sum _{j=1}^9 \alpha _j (a_j-a_{j-1}) =1\) to ensure that \(\phi (u,\alpha )\) is a proper density function.
The proposed back-calculation method estimates a total of \({\hat{N}}\)=13,130 individuals (standard error [SE] 15; 95% confidence interval [CI] 13,101–13,159) who were infected before March 15. This includes 13,071 confirmed cases on and before March 15, which means that we expect 59 = 13,130-13,071=\({\widehat{N}}-n\) additional cases to be confirmed after March 15, should there be no new infections. The expected numbers of new cases under the fitted model are shown in Fig. 2a. The predicted numbers track well with the observed data, suggesting that the proposed model fits well. The maximum likelihood estimates of the shape and size parameters for the Weibull distribution are 1.03 (SE 0.004) and 5.39 (SE 0.17), respectively. This corresponds to a median of 3.8 days (SE 0.12). As discussed before, the incubation distribution is estimated subject to a location shift. Recently, Qin et al. [12, p. 11] analyzed the length-biased incubation time from 1211 confirmed COVID-19 cases who left Wuhan before the lockdown and reported a median incubation period of 8.13 days (95% CI 7.37–8.91). This suggests a location shift factor of about 5 days in our estimates.
What’s also of interest is the infection density estimate after the lockdown of Wuhan. The maximum likelihood estimates for \(a_j\)’s in the 9 selected time intervals, including the time interval preceding January 20, are \((2.56\times 10^{-5}, 5.03\times 10^{-2}, 7.34\times 10^{-2}, 1.74\times 10^{-2}, 7.72\times 10^{-12}, 3.55\times 10^{-14}, 6.75\times 10^{-5}, 7.92\times 10^{-4}, 9.05\times 10^{-4})\) with standard errors \((1.08\times 10^{-5}, 1.53\times 10^{-3}, 1.29\times 10^{-3}, 1.40\times 10^{-3}, 4.93\times 10^{-4}, 1.07\times 10^{-11}, 1.21\times 10^{-4}, 1.77\times 10^{-4}, 2.64\times 10^{-4}\)). By definition, these estimates are for infected individuals who have been or will be diagnosed. Since the number of diagnosed individuals is proportional to the total number of infected (including those who were asymptomatic), the values provide estimates of relative risk among all infected individuals. After accounting for a location shift factor of 5 days, the peak of infection occurred immediately after the lockdown of Wuhan city (Fig. 2b). Moreover, the incidence increases 46% during the week of Wuhan lockdown from the preceding week, but dropped 76% in the next week, suggesting that the travel ban that started from Wuhan and its neighboring cities had initially increased the disease spread in other provinces, but was able to effectively slow down the spread of the disease in just two weeks.
Finally, continuous assessment of potential new cases to be diagnosed in the near future can be implemented by applying the back-calculation algorithm using up-to-date data. Figure 2c shows the projected number of confirmed cases with data available up to that time point, assuming no new infections afterward. The difference between the expected and observed cumulative number of confirmed cases gives the near-term prediction of additional cases to be diagnosed. Since the prediction is relatively unstable for the first few weeks of the epidemic due to the limited amount of data, we only perform prediction after February 10, that is, the fourth week into the epidemic. As shown in Fig. 2c, the prediction obtained after February 24 is very close to the total number of confirmed cases at the end of the study period (March 15). Ignoring the spike occurred around February 20, which was most likely caused by the delayed report of cases in two prisons, the prediction algorithm performs reasonably well after February 17. This shows that the back-calculation algorithm can potentially provide a useful utility for the planning of health care allocation, especially for an epidemic that is still growing.
COVID-19 epidemics in Hubei province, China
We also apply the proposed back-calculation algorithm to evaluate the COVID-19 epidemics in Hubei province. As in the previous section, we focus on data reported during the 8-week period between January 20 and March 15. Note that the diagnosis classification rule was relaxed for cases in Hubei province after February 12, 2020. Among the 14,840 confirmed cases reported on February 12, 13,332 cases did not meet the old diagnosis rule. The exceptionally large number 4823 of new cases on February 13 may also include cases that did not meet the old diagnosis rule. To reduce the impact caused by the modification of the classification rule, we set the numbers of new cases on February 12 and 13 under the old classification rule to be equal and both to \(14,840-13,332 = 1508\). Then we redistributed the extra \(13,332+4823-1508 = 16,647\) cases to each of the 25 days preceding February 14 according to the number of reported cases under the old classification rule.
Applying the modified back-calculation procedure, we estimated that 68,617 individuals (SE 13; 95% CI 68,592–68,642) were infected before March 15 in the Hubei province. Since a total of 68,499 confirmed cases were reported on and before March 15, suggesting that at least 118 additional cases will be confirmed after March 15. The observed, adjusted, and expected numbers of new cases are shown in Fig. 3a. Again, the proposed model fits well as the predicted epidemic sizes are very close to the observed ones.
The maximum likelihood estimates of the shape and size parameters for the imposed Weibull distribution are 1.02 (SE 0.001) and 6.98 (SE 0.096), respectively. This corresponds to a median of 4.9 days (SE 0.10). Compared with the median incubation time of 8.13 days (95% CI 7.37–8.91) reported in Qin et al. [12, Page 11], this suggests a location shift factor of approximated 4 days in our model fit. Moreover, the estimated infection density in the Hubei province (see Fig. 3B) exhibits a similar pattern as that for regions outside the Hubei province. The risk of infection was the highest during the 3-week period between January 20 and February 10, with estimated intensity of 0.02 (SE \(4\times 10^{-4}\)), 0.08 (SE \(8\times 10^{-4}\)), and 0.04 (SE \(8\times 10^{-4}\)) in these three consecutive weeks. This implies that the incidence increases 300% during the week of Wuhan lockdown from the preceding week, but dropped 50% in the next week. Again this result supports that the strict travel ban initially increased the disease spread, but later slowed down the spread of the disease in a short time.
COVID-19 epidemics in South Korea
Due to the soaring COVID-19 cases, South Korea raised the alert level to the highest, Red, on February 23 and quickly adopted a “test, trace, isolate, and treat” strategy to contain the spread of the virus. To further illustrate the proposed method, we now analyze data from South Korea during the period between February 20 and April 20, 2020. The cutoff date was selected because restrictions on social distance were relaxed after April 20, which can potentially lead to a remarkable increase in the number of new confirmed cases. We set \(\tau _0\) as February 13, 2020 to account for the possible infection period. The proposed method gives \({\hat{N}} =\)10,682 (SE 11; 95% CI 10,661–10,703), and projects at least \(X_{K+1} = 39\) additional cases to be confirmed after April 20, 2020. The lower panel of Figure 3 shows the observed and expected numbers of daily new confirmed cases, as well as the estimated infection density.
The maximum likelihood estimates of the shape and size parameters for the Weibull distribution are 0.87 (SE 0.02) and 5.39 (SE 0.25), respectively, corresponding o a median of 1.78 days (SE 0.15) in days to diagnosis. The highest infection risk occurred during the two-week interval between February 20 and March 5, where the estimated intensity in these two weeks are 0.0264 and 0.0775. The risk stayed low after March 5, with an estimated intensity lower than 0.01. To the best of our knowledge, we are not aware of any published data on the incubation distribution in South Korea. However, if the same reference number as in China is used, we estimate the location shift parameter to be approximately 6. After accounting for the location shift factor, the peak of infection occurred again immediately after the prevention and control strategy took place in South Korea. This indicates that the prevention and control strategy adopted by the South Korea government had initially increased the disease spread, but was able to have it slowed down remarkably in 2 weeks.
Conclusion
Different countries have taken different measures in response to the novel coronavirus, and there has been a continuing heated debate on whether aggressive COVID-19 control measures cost more than they are worth. Among all countries, China has imposed the most sweeping restrictions in response to COVID-19. The authorities locked down cities, restricted movements of millions and suspended business operations to prevent further outbreak of the disease. South Korea, another country on the front-line of the epidemic, adopted the “test and trace” strategy to aggressively test people for the disease and quarantine those who tested positive, so that the rest of the population can go about their daily lives. Both countries have observed a significant decline in the number of confirmed cases. Our analysis provides some preliminary evidence on the effectiveness of these measures by analyzing data from China and Korea. We conclude that the extreme measures undertaken by the Chinese government and the “test and trace” strategy adopted by Korea government have effectively slowed down the spread of the disease in both countries in about two weeks.
Discussion
In this paper, we propose to modify the back-calculation algorithm by imposing a parametric model for the incubation distribution, thus avoid the requirement of a known incubation distribution. Such an extension, however, induces an identifiability issue, that is, the incubation distribution and the infection density are identifiable up to a location shift. In the case where the incubation time is of interest, we suggest to approximate the location shift factor by comparing the estimated mean/median incubation time to what reported in the existing literature. We also demonstrate that the back-calculation algorithm can be used to estimate the number of infected individuals to be diagnosed in the near future. This provides a useful utility to guide the planning of medical resource allocation in the middle of the epidemic.
As mentioned in Section 2, the distribution of the incubation time, that is the time from infection to diagnosis, depends on the testing strategy and thus may evolve over time due to policy change. Hence it is desirable to allow the incubation distribution to be time-dependent. However, in this case, model fitting may be unstable and requires external information to guide the selection of the parametric model form. Future research on developing a stable algorithm that can account for the time-dependent incubation distribution is warranted.
Data availability
The data are available from the COVID-19 Data Repository by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University.
Code availability
The R codes used in this paper are available on request.
References
Li Q, Guan X, Wu P, et al. Early transmission dynamics in Wuhan, China, of novel coronavirus-infected pneumonia. N Engl J Med. 2020;. https://doi.org/10.1056/NEJMoa2001316.
Imai N, Cori A, Dorigatti I, et al. Report 3: Transmissibility of 2019-nCoV. 2020; www.imperial.ac.uk/mrcglobal-infectious-disease-analysis/news--wuhan-coronavirus/.
Read JM, Bridgen JR, Cummings DA, et al. Novel coronavirus 2019-nCoV: early estimation of epidemiological parameters and epidemic predictions. medRxiv. 2020; https://doi.org/10.1101/2020.01.23.20018549
Zhao S, Lin Q, Ran J, et al. Preliminary estimation of the basic reproduction number of novel coronavirus (2019-nCoV) in China, from 2019 to 2020: a data-driven analysis in the early phase of the outbreak. Int J Infect Dis 2020;92:214–7.
Report of the WHO-China Joint Mission on Coronavirus Disease 2019 (COVID-19) - World Health Organization, Feb. 28, 2020. https://www.who.int/docs/default-source/coronaviruse/who-china-joint-mission-on-covid-19-final-report.pdf
McEvoy M, Tillet HE. Some problems in the prediction of future numbers of cases of the acquired immunodeficiency syndrome in the U.K. Lancet. 1985;2:541–2.
Morgan WM, Curran JW. Acquired immunodeficiency syndrome: current and future trends. Public Health Rep. 1986;101:459–64.
Bailey N. The mathematical theory of infectious diseases. 2nd ed. London: Charles Griffin & Co.; 1975.
Brookmeyer R, Gail MH. Minimum size of the acquired immunodeficiency syndrome (AIDS) epidemic in the United States. Lancet. 1986;328(8519):1320–2.
Brookmeyer R, Gail MH. A method for obtaining short-term projections and lower bounds on the size of the AIDS epidemic. J Am Stat Assoc. 1988;83(402):301–8.
Ferguson NM, Donnelly CA, Woolhouse MEJ, et al. The epidemiology of BSE in cattle herds in Great Britain. II. Model construction and analysis of transmission dynamics. Philos Trans R Soc Lond Ser B. 1997;352(1355):803–38.
Qin J, You C, Lin Q, et al. Estimation of incubation period distribution of COVID-19 using disease onset forward time: a novel cross-sectional and forward follow-up study. medRxiv. 2020; https://doi.org/10.1101/2020.03.06.20032417.
Acknowledgements
We are deeply grateful to Professor Jinglong Wang for his help and encouragement in the process of completing this manuscript. We also thank the editor and two anonymous referees for their valuable feedback and suggestions that significantly improved the manuscript. Yukun Liu was supported by the National Natural Science Foundation of China (Grant No. 11771144), the State Key Program of National Natural Science Foundation of China (Grant No. 71931004), Natural Science Foundation of Shanghai (Grant No. Grant No. 19ZR1420900), the development fund for Shanghai talents, and the 111 project (Grant No. B14019). Yan Fan was supported by the National Natural Science Foundation of China (Grant No. 11971300) and Natural Science Foundation of Shanghai (Grant No. 17ZR1409000). Yong Zhou research was supported by the State Key Program of National Natural Science Foundation of China (Grant No. 71931004), and the State Key Program in the Major Research Plan of National Natural Science Foundation of China (Grant No. 91546202).
Funding
Yukun Liu was supported by the National Natural Science Foundation of China (Grant No. 11771144), the State Key Program of National Natural Science Foundation of China (Grant No. 71931004), Natural Science Foundation of Shanghai (Grant No. 19ZR1420900), the development fund for Shanghai talents, the 111 project (Grant No. Grant No. B14019), and the Fundamental Research Funds for the Central Universities. Yan Fan was supported by the National Natural Science Foundation of China (Grant No. 11971300) and Natural Science Foundation of Shanghai (Grant No. 17ZR1409000). Yong Zhou research was supported by the State Key Program of National Natural Science Foundation of China (Grant No. 71931004), and the State Key Program in the Major Research Plan of National Natural Science Foundation of China (Grant No. 91546202). The content is solely the responsibility of the authors and does not necessarily represent the official views of the funding agencies.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Liu, Y., Qin, J., Fan, Y. et al. Estimation of infection density and epidemic size of COVID-19 using the back-calculation algorithm. Health Inf Sci Syst 8, 28 (2020). https://doi.org/10.1007/s13755-020-00122-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13755-020-00122-8