Abstract
Digital image processing techniques are being explored for accurate and timely diagnosis of malaria, a serious parasitic infection of humans. A key decision factor in the diagnosis is the degree of infection, also called parasitaemia. This paper presents an efficient method for estimating parasitaemia using the digital images of thin blood smears that has been stained with Gimsa or equivalent stain. The method utilizes the 4-connected set properties of digital images to identify the various regions existing within the image. Properties of the different identified regions, such as centroids, major and minor axis, etc., are used to arrive at the number of RBCs (good and infected) present in the image. The method addresses the issues of partially visible RBCs, as well as that of overlapped RBCs. It also addresses image imperfections, caused by dust on the slide, etc.
Similar content being viewed by others
1 Introduction
Malaria is caused by protozoan parasites of the genus plasmodium and is transmitted by the bite of anopheles mosquito. Four species of the plasmodium parasite infect humans: P. falciparum, P. vivax, P. ovale and P. malariae. The parasite’s lifecycle within humans can be divided into three distinct stages: Trophozoite, Schizont, and Gametocyte. During this lifecycle, human red blood cells (RBCs) are used as host. The shape and size of the parasite differs by the species and lifecycle stage of the parasite. However in each lifecycle stage the parasite has at least one chromatin, which is the nucleus of the parasite. A measure of severity of the infection, called parasitaemia, is the ratio of the parasite infected RBCs to the total number of RBCs. This is an important determinant in selecting appropriate treatment and drug dose.
Currently clinical diagnosis utilizes microscopy to study the prepared blood smears. However this is extremely time consuming and is dependent on the skill and experience of the examiner, and hence has limited reliability. Thus it is important to develop an automated image analysis system that can identify and count infected and un-infected RBCs in the images of blood smears. Further, in the clinical process, two different types of blood smears (thick and thin) are produced. The thin smear is used to identify the type of parasite and the thick smear to estimate parasitaemia. However, the complete information is available in the thin smear.
In this paper we present a technique for estimating parasitaemia in the images of stained thin smears of blood. The technique presented is computationally efficient and fast and automatically adapts to the variations in images such as magnification, object orientations, etc. It also addresses challenges presented by presence of dust or leftover stain on the slide. The method utilizes 4-connected sets to identify different regions existing in the image. Properties of these regions, such as area, coordinates of centroids, major and minor axes and Euler’s numbers are utilized to take decisions. Key challenges to the counting process are : 1) partially visible cells at the boundary of the image and 2) overlapped cells. The image is first processed to identify the presence of any external artifacts and address them. The total number of cells present in the field of view is then estimated. Then the number of infected cells is calculated. Using this data the degree of parasitaemia can be calculated.
The technique however assumes that the presence of the parasite in the image has already been established by the use of other automated techniques [5–9]. Further, the image presented for analysis should be a true colour RGB image captured with adequate magnification and resolution. It should be noted that this method cannot be applied to images of thick smears.
2 Literature Review
Most of the available literature concentrates on segmenting the chromatin dots within the RBC [5–11], to establish the presence of the parasite. Of the available literature, [3, 4] concentrate on measuring parasitaemia. Of these [4] uses an image mosaicing system to study the partially visible cells at the edge of the FoV. This may not be realizable in a practical clinical situation, and hence has limited utility. The method described in [3] uses a synthesized template of an RBC, parameterized using variable eccentricity and major axis. The shape of the RBC is assumed to be elliptical. This template matching process gives less than acceptable results for overlapped cells, partially visible cells at the edge of the FoV, and when the cells are oriented at an angle, resulting in poor matching with the template. Besides the method used to arrive at the radius of the RBC is very slow.
Hence there is need for method that is not affected by shape, size, orientation, or location of the RBC in the FoV.
3 Methodology
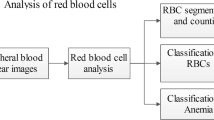
The analysis process consists of six distinct steps (Fig. 1). In the first step the image is studied for appropriateness for this study. The image is expected to be a true colour RGB image. Besides, the smear should have been stained with a gimsa or equivalent stain. This stain renders specific colour to specific portion of the parasite [2], irrespective of the lifecycle stage of the parasite: chromatin stains to a deep red colour, cytoplasm stains to a shade of blue but the exact colour varies from species to species, RBCs become pale-yellowish pink, vacuoles do not take any colour. To verify that the smear has been stained with appropriate stain, a copy of the image is converted to HSI colour space, and a population of pixels in the six hue ranges indicated in Table 1 is taken. An image is accepted for analysis only if it exhibits population in red and blue ranges. The entire hue range of 0–3600 is divided into six segments with each segment having a width of 600, and centered on a colour indicated in Table 1. The range of hues associated with each colour takes care of variances in colour existing in different images.
This first step is however required only of this system is used as a standalone system. This step is not required if this process is integrated with other system for detecting the presence of malaria.
A candidate image identified by the previous step is then converted to a gray scale image. This image is used to count RBCs.
Figure 2 show extracts from images of thin smears of blood that has been stained to display malaria. These are gray version of the original true colour images. This set would be used to demonstrate this technique. Dataset 1 is used to explain the methodology too.
The gray image is then converted to a binary image by thresholding it. To do this, we utilize the fact that the image has a predominant background that has intensity distinct from that of the foreground. The histogram is thus bi-modal (Fig. 3a). Otsu’s method was thus used to arrive at the threshold value. This was used to create the binary image shown in Fig. 3b.
The binary image shows two types of imperfections : 1) artifacts in the background region (caused by dust, foreign bodies in slide, etc.), and 2) holes within the RBCs. Both these issues need to be addressed before proceeding. Similar procedure is used to address both the issues. We address issue 1 before addressing issue 2. Essentially, the 4-connected set property of digital images would be used to identify various regions existing in the image. The area of the regions would be used to take decisions. The study is done on a copy of the binary image. When some pixels are identified for correction, the correction is done in the original binary image.
The artifacts in the background region are black objects on a white background. To remove the artifacts, a digital negative of the copy of the binary image is converted into a 4-connected region labeled image. The population of each labeled region is an indication of area occupied by each region. This population data is used to identify these small artifacts. Any region with area less than 0.3 % of the image size was erased and the corresponding pixels in the binary image were marked as background pixels. This also removed some very small partially visible cells at the boundary of the image. Since they are not counted, this is not an issue. Figure 4a.
Unlike the artifacts in the background, the holes within the RBCs are white objects in black background, and hence, the corrected binary image was converted into a 4-connected region labeled image again. As before, population of each labeled region was calculated and the regions with population less than particular threshold were marked as foreground pixels in the binary image. The threshold value is essentially the area of a circle of radius equal to 60 % of the radius of an RBC. (Fig. 4b). The method used to measure the radius of the RBCs is described later.
The corrected binary image will now be used to count the number of RBCs present in it. The RBCs are essentially visible as different objects in the field of view (FoV). The objects visible can be categorized into two groups: 1) partially visible RBCs existing at the edges of the FoV, and 2) Objects within the FoV. The second group consists of free standing RBCs and overlapped RBCs. These three categories of features can be easily differentiated once the image is converted into a 4-connected region labeled array. Features of the disjoint regions, such as coordinates of centroids and lengths of major and minor axis can be used to arrive at decisions. The data for the regions in data set 1 are shown in Table 2.
The first step in the counting process is to determine the diameter of the RBCs. This would vary from image to image, depending on the magnification used to capture the image. Within an image too some variation is expected due to infection or orientation of the RBC at the time of image capture. The ratio of the major axis to minor axis of all the identified regions are studied. Regions having this ratio ~1 represent free-standing RBCs. The minor axis of a region with minimum ratio (≈1 and >1) is taken as the diameter of the RBC, in pixels. The measure is rounded to the nearest integer.
The next step is to count the partially visible cells at the edge of the FoV. These cells have their centroids at a distance less than the diameter of a RBC, from the edge of the image. Distance of the centroid from the four edges of the image is considered for this decision. The number of partially visible cells is then counted. Partially visible cells can also be overlapped. For such cells the ratio of major to minor axis is >1. To calculate the number of overlapped RBCs the ratio of the major axis to the diameter of an RBC is used for decisions. This data is used to correct the count of partially visible cells. This resolution is however done only if the minor axis is more than 80 % of the diameter of a RBC.
The next step is to count the number of free standing RBCs. As discussed before such cell have the ratio of their major to minor axis ~1, and are located within the FoV.
The partially overlapped RBCs are identified by regions having their ratio of major to minor axis greater than a threshold value. For such regions the ratio of the major and minor axis to the diameter of an RBC is used to arrive at the number of overlapped RBCs in the region.
The sum of the count of the free standing cells, partially visible cells and overlapped cells gives the count of RBCs present in the image.
To count the infected RBCs, we need an image with the chromatin dots segmented. This can be obtained following any of the methods described in [5–11]. Figure 6 shows an image generated following one such method. A digital negative of this image is added to the image in Fig 4(b), and 4-connected set data and region properties regenerated. Eulers numbers of the regions indicates the regions that have another region embedded within it, i.e., the infected RBCs.
4 Experimental Results
The prototype was developed using MATLAB. The developed algorithm was tested using images sourced from Center for Disease Control and Prevention’s malaria image library [1].
For generating the binary mask Otsu’s method was used to generate the threshold. For image in Fig. 2(a) Otsu’s formula returned a threshold value of 227. The histogram is shown in Fig. 3(a), and the segmented image in Fig. 3(b). The segmented image shows some small external artifacts in the background region.
On converting a digital negative of the image to a 4-connected region labeled image we get 68 regions of which 37 have a population less than the threshold value of 270 for this image (0.3 % of image area of 300 × 300). These regions are removed from the binary mask, see Fig. 4(b).
The process described was able to remove artifacts from the background region when any external object on the slide result in free standing objects in the background region in the binary image. However, artifacts that result in objects that are attached to other RBCs are not removed.
The cell counting process is dependent on the existence of at least one free standing RBC in the FoV. This is expected to exist in images of thin smears. However, in the extreme case when no such RBC exists, the process would fail.
After correcting for imperfections, dataset 1 had 30 4-connected regions. The properties of these regions are shown in Table 2.
Diameter of a free standing cell (identified by ratio ~1) is the diameter of the RBC. Here, diameter of free standing cell = 36. The region taken for measuring the diameter was region 17, which had the minimum ratio (1.02). While measuring the diameter, care should be taken that the target region is completely located within the FoV. Thus after measuring the diameter, the distance of the centroid of the target region from the edges is measured, to ensure that it is at a distance greater than the diameter from the edges.
Partially visible objects, whose centroids are within a distance of unit radius of an RBC from the boundary of the image, are represented by regions 1,6,12,25,29,30. Table 3 is an extract from Table 2, and shows the properties of these regions. Of these, region 12 represents two overlapping cells. For the other cases, even though the ratio was high, the minor axis is less than 80 % of the diameter of an RBC, and hence was not resolved.
Partially visible cells = 7
Twenty regions had the ratio less than the threshold value of 1.3 and were treated as free standing cells. Region 20 had high ratio, but the minor axis was 77 % of the diameter (<80 % threshold) and hence was treated as free standing cell.
Free standing cells = 21
Partially overlapped cells are identified by their ratio being greater than a threshold value. The application currently uses a threshold value of 1.3. For such cases the major axis and minor axis is divided by the diameter and rounded to the nearest integer to arrive at the number of overlapped cell. This resolution is not done for cases where the minor axis is less than 80 % of the diameter of the RBC. Regions 13,19 and 24 qualified as overlapped regions and were resolved. Table 5 shows properties of such regions.
The process worked fairly accurately for partially overlapped cells involving two RBCs. However when more than three RBCs were involved the degree of success dependent on the degree of overlap. However, trained technicians are expected to produce slides with minimum overlapped RBCs.
5 Conclusion
Using the algorithm described, we were able to successfully demonstrate that it is possible to build an automated system to measure parasitaemia in the images of thin blood smears.
References
Center for Disease Control and Prevention-malaria image library, available at : http://www.dpd.cdc.gov/dpdx/HTML/ImageLibrary/Malaria_il.htm. Accessed July 2013
Medical Chemical Corporation, Giemsa stain: Pre-analytical considerations. http://www.med-chem.com/procedures/Giemsabsp.pdf. Accessed Feb 2011]
Halim S, Bretschneider T, Li Y, Preiser P, Kuss C (2006) Estimating malaria parasitaemia from blood smear images. In: Proceedings ICCARV IEEE, p 648–653
Zou L-H, Chen J, Zhang J, Garcia N (2010) Malaria cell counting diagnosis within large field of view, digital image computing: techniques and applications (DICTA), 2010 international conference on, vol., no., pp.172–177 1–3 Dec 2010
Di Ruberto Cecilia, Dempster Andrew, Khan Shahid, Jarra Bill (2001) “Morphological Image Processing for Evaluating Malaria Disease”, IWVF4. LNCS 2059:739–748
Dempster AG, Di Ruberto C (2001) Using granulometries in processing images of malarial blood. In: Proceedings of the international symposium on circuit and systems, p 291–294
Di Ruberto C, Dempster AG, Khan S, Jarra B (2000) Segmentation of blood images using morphological operators. Proc Int Conf Pattern Recognit 3:397–400
Elmoataz A, Revenu M, Porquet C (1992) Segmentation and classification of various types of cells in cytological images. In: Proceedings of the international conference on image processing and its applications, p 385–388
Ghosh S, Ghosh A (2014) Content based retrieval of malaria positive images from a clinical database via recognition in RGB colour space. In: Proceedings of 48th annual convention of CSI–volume II, advances in intelligent systems and computing 249, p 1–8
Ghosh S, Ghosh A (2014) Content based retrieval of malaria positive images form a clinical database. In: Proceedings of the 2013 IEEE second international conference on image information processing, p 313–318
Ross NE, Pritchard CJ, Rubin DM, Dus AG (2006) Automated image processing method for the diagnosis and classification of malaria on thin blood smears. Med Biol Eng Comput 44(5):427–436
Acknowledgments
Financial assistance from the Department of Applied Optics and Photonics, University of Calcutta, under the Technical Education Quality Improvement Program is acknowledged by the first author. Opinions expressed and conclusions drawn are of the authors and are not to be attributed to the Department of Applied Optics and Photonics.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ghosh, S., Ghosh, A. & Kundu, S. Estimating malaria parasitaemia in images of thin smear of human blood. CSIT 2, 43–48 (2014). https://doi.org/10.1007/s40012-014-0043-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40012-014-0043-7