Abstract
A kind of spectral meshless radial point interpolation method is proposed to degenerate parabolic equations arising from the spatial diffusion of biological populations and satisfactory agreements is archived. This method is based on collocation methods with mesh-free techniques as a background. In contrast to the finite-element method and those meshless methods based on Galerkin weak form, such as element-free Galerkin, there is no integration tools in this approach. Furthermore, some numerical experiments are given to validate the accuracy of the results and stability of the present method.
Similar content being viewed by others
Introduction
The biological population problems have attracted much attention and research recently [1, 2]. Biologists believe that dispersal or emigration play a key role in the regulation of population of some species. A persuasive example of this suggestion occurs in a paper by Carl [3], whose observations on a population of arctic ground squirrels showed that this species achieves population control by the dispersal of animals from densely populated areas of favorable habitat into unfavorable areas, where burrow sites are not available, and where they are subjected to intensive predation.
Consider the following nonlinear biological population model
with a given initial condition \(u({\mathbf{x}} ,0)\), where u denotes the population density, and \(\sigma\) represents the population supply due to births and deaths. We can consider a more general form for \(\sigma\) as \(hu^a(1-ru^b)\), in which \(h, a, r, b\in \mathbb {R}\). The field \(u({\mathbf{x}} ,t)\) gives the number of individuals’ per-unit volume at position \({\mathbf{x}}\) and time t, and it is integral over any subregion \(\Omega\) to give the total population of \(\Omega\) at time t. The field \(\sigma (u)\) gives the rate at which individuals are supplied (per-unit volume) exactly at \({\mathbf{x}}\) through the births and deaths. The flow of population from point to point is then depicted by the diffusion velocity \(v({\mathbf{x}} ,t)\), which provides the average velocity of those individuals who occupies \({\mathbf{x}}\) at the time t. The fields u, v, and \(\sigma\) should be consistent with the following law of population balance (for any regular subregion \(\Omega\) for the defined region during all the time t):
where n is the outward unit normal vector to the boundary \(\partial \Omega\) of \(\Omega\). This equation means that the rate of change of population of \(\Omega\) addition to the rate at which individuals left \(\Omega\) across its boundary should be equal to the rate at which individuals are directly supplied to \(\Omega\) [4, 5].
Several papers have considered the existence, uniqueness, and regularity of weak solutions [4–9] for general degenerate parabolic equations of Eq. (1). Numerical solutions of the biological population equation have seldom been explored and investigated, though some numerical struggles have been done in this field. Modeling of the biological population problem (1) has been explored using the element-free Kp-Ritz method in [1]. An improved element-free Galerkin method for numerical modeling of the biological population problem (1) has also been applied by Zhang et. al. [2]. Shakeri and Dehghan obtained numerical solution of the model using He’s variational iteration method [10].
In general, the mesh-free methods can be grouped into two categories, first category uses the equation in weak form, for instance, element-free Galerkin method (EFG), improved element-free Galerkin method (IEFG), complex variable element-free Galerkin method (CVEFG), improved complex variable element-free Galerkin method (ICVEFG), meshless local Petrov–Galerkin method, and etc. [11–21], and the second category applies the strong form of the equation, for example, meshless collocation approach [22–28]. In addition, the meshless methods depend on how their shape or basis function constructed are different, and it might be based on interpolation or curve fitting and etc. [29–41].
In the current work, we testify spectral meshless radial point interpolation (SMRPI) method [42–44] on the problem (1) and then make simulations on two numerical experiments which leads to satisfactory results. A technique based on radial point interpolation is adopted to construct shape functions, also called basis functions, using radial basis functions. These shape functions have delta function property in the frame work of interpolation; therefore, they convince us to impose boundary conditions directly. The time derivatives are approximated by the finite-difference time-stepping method. This method is based on collocation methods with mesh-free techniques as a background. In contrast to those meshless methods based on weak form, there is no integration tools in this approach. Therefore, the computational complexity of SMRPI method seems to be of low order.
The time discretization approximation
In the current work, let us to employ a time-stepping scheme to evaluate the time derivative. To this end, the following first-order finite-difference approximation for the time derivative operator is adopted:
Moreover, we apply the following approximations using the Crank–Nicolson technique:
where \(u^{k}({\mathbf{x}} )=u({\mathbf{x}} ,k\Delta t)\) and \({\mathbf{x}} =(x,y)\). Applying the above approximation and impose them to the original Eq. (1), we are conducted to the following time discretization equation:
The basis functions in the frame of MLRPI
Consider a continuous function \(u({\mathbf{x}} )\) defined in a domain \(\Omega\), which is represented by a set of field nodes. The \(u({\mathbf{x}} )\) at a point of interest \({\mathbf{x}}\) is approximated in the form of
where \(R_i({\mathbf{x}} )\) is a radial basis function (RBF), n is the number of RBFs, \(p_j({\mathbf{x}} )\) is monomial in the space coordinate \({\mathbf{x}}\), and m is the number of polynomial basis functions. Coefficients \(a_i\) and \(b_j\) are unknown which should be determined. In the current work, we use the thin plate spline (TPS) as radial basis functions in Eq. (7) which is defined as follows:
To determine \(a_i\) and \(b_j\) in Eq. (7), we consider a support domain, such as a disk or square, surrounding the point of interest \({\mathbf{x}}\) and use all nodes included in the support domain to form a system of equations based on Eq. (7). In this way, coefficients of \(a_i\) and \(b_j\) are obtained. Therefore, by the idea of interpolation, Eq. (7) is converted to the following form:
where \(\phi _i({\mathbf{x}} )\) are called the RPIM shape functions which have the Kronecker delta function property, that is
This is because the RPIM shape functions are created to pass thorough nodal values. Moreover, the shape functions are the partitions of unity, that is
for more details about RPIM shape functions and the way they are constructed, the readers are referred to see [42].
Operational matrices of high-order derivatives
The essential tools of the current approach is operational matrices which is constructed and described briefly in this section. In fact, the operational matrices make the method easy to apply the high-order differential equations which are really difficult to handle by the majority of methods, especially those techniques based on weak forms. Consider the total N nodes for covering the domain of the problem, i.e., \(\overline{\Omega }=\left( \Omega \cup \partial \Omega \right)\). On the other hand, as we noted in the previous section, n is depend on the point of interest \({\mathbf{x}}\) (so, we call it \(n_{\mathbf{x}}\)) in Eq. (9) which is the number of nodes included in support domain \(\Omega _{\mathbf{x}}\) corresponding to the point of interest \({\mathbf{x}}\). Therefore, we have \(n_{\mathbf{x}}\le N\) and Eq. (9) could be reformulated as
In fact, there is one shape (basis) function \(\phi _j({\mathbf{x}} )\), \(j=1,2,3,{\ldots },N\) corresponding to the node \({\mathbf{x}}\), we define \(\Omega _{\mathbf{x}}^c=\left\{ {\mathbf{x}}_j:{\mathbf{x}} _j\notin \Omega _{\mathbf{x}}\right\}\), and then, it is straightforward, from the previous section, to conclude
The derivatives of \(u({\mathbf{x}} )\) are easily obtained as
and also high derivatives of \(u({\mathbf{x}} )\) are clearly obtained as
where \(\frac{\partial ^{s}(\cdot )}{\partial x^s}\) and \(\frac{\partial ^{s}(\cdot )}{\partial y^s}\) are the sth derivative with respect to x and y, respectively. By indicating \(u_x^{(s)}(\cdot )=\frac{\partial ^{s}(\cdot )}{\partial x^s}\) and \(u_y^{(s)}(\cdot )=\frac{\partial ^{s}(\cdot )}{\partial y^s}\), and substituting \({\mathbf{x}} ={\mathbf{x}} _i\) in Eq. (14), we can formulate the discrete differentiations process as a matrix-vector multiplications
we indicate above coefficients matrices as \({\mathbf{D}}x\) and \({\mathbf{D}}y\), respectively. In addition, clearly, we propose the following matrix-vector multiplications for high-order derivatives
where
Discretization and numerical implementation for the method
Equation (6) can be rewritten as
To overcome nonlinearity, suppose u k + 1 ≊ u k in the right-hand side of the above equation. This is possible if the time step \(\Delta t\) be sufficiently small, therefore, Eq. (24) can be converted to
Now, consider N scattered nodes on the boundary and domain of the problem (1), i.e., \(\overline{\Omega }=(\Omega \cup \partial \Omega )\). Assuming that \(u({\mathbf{x}} _i,k\Delta t),\ i= 1,2,{\ldots },N\) are known, our purpose is to compute \(u({\mathbf{x}} _i,(k+1)\Delta t),\ i= 1,2,{\ldots },N\). For nodes which are included in the inside of the domain, i.e., \({\mathbf{x}} _i \in \Omega\), to obtain the discrete system of algebraic equations, let us substitute approximate formulas (12) and (14), (15) into equation (25), then
Now, by setting \({\mathbf{x}} ={\mathbf{x}} _i\), \(i=1,2,3,{\ldots },N_\Omega\) (\(N_\Omega\) is the number of nodes in \(\Omega\)) in the above equation and then applying notations (19)–(23), we obtain
Enforcement of boundary conditions
There are several techniques to enforcing essential boundary conditions in meshless methods, such as the use of penalty methods, Lagrange multipliers and modified variational principles, etc. In the current work, the essential boundary conditions are imposed directly.
Simply-supported boundary conditions
In the case of simply-supported boundary conditions, we have
For nodes which are located on the boundary \(\partial \Omega\), we set as
where \(N_{\partial \Omega }\) is the number of nodes located on \(\partial \Omega\).
Clamped boundary conditions
In the case of clamped boundary conditions, it is usually included the following types of boundary conditions:
Therefore, in this case, for nodes which are located on the boundary \(\partial \Omega\), we set as
where \({\mathbf{n}} ={\mathbf{n}} _1({\mathbf{x}} _i)i+{\mathbf{n}} _2({\mathbf{x}} _i)j\) is the outward unit normal to the boundary \(\partial \Omega\) at \({\mathbf{x}} _i\in \partial \Omega\). Then, we have the following equations:
where \(N_{\partial \Omega }\) is the number of nodes on \(\partial \Omega\).
Numerical simulations
In this section, we aim to demonstrate that the SMRPI approach has a wider applications by testifying two examples of the type in Eq. (1). To show the accuracy and convergence of the method, maximum absolute error \(\varepsilon _{\infty }\) defined by
is used, where \(\lbrace u_\mathrm{{approx}}, p_\mathrm{{approx}}\rbrace\) are exact and numerical SMRPI solutions, respectively. In implementing the SMRPI method, we consider support domain as a disk with the radius \(r_s= 4.2h\), where \(h=0.1\) is the distance length between two nodes in x- or y-directions.
Example 1
Considering the following biological population equation:
with the initial condition
with \(h=\frac{1}{5}\), \(a=1\), and \(r=0\), and the exact solution is
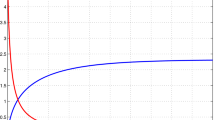
In this problem, we set \(\Delta t = 0.00001\). Figure 1 shows SMRPI simulations and the absolute errors in some levels of specific values of the time. Figure 2 compares the exact and approximate SMRPI solutions.
Example 2
Considering the following biological population equation:
with the initial condition
with \(h=-1\), \(a=1\), \(r=-\frac{8}{9}\), and \(b=1\), and the exact solution is
In this problem, we set \(\Delta t = 0.00001\) as well. Figure 3 shows SMRPI simulations and the absolute errors in some levels of specific values of the time. Figure 4 compares the exact and approximate SMRPI solutions. As it is seen, the SMRPI and the exact solutions are not distinguishable, while we have adopted a very simple idea to overcome the nonlinearity, as it has been pointed out in Sect. "Discretization and numerical implementation for the method".
Conclusions
In this paper, the biological population equation has been investigated using the spectral meshless radial point interpolation method. The shape (basis) functions constructed by radial point interpolation augmented to monomials have been employed to approximate the 2D displacement field. A system of nonlinear discrete equations is obtained through application of the SMRPI. The nonlinear equation system is solved by iteration with a very simple scheme. A mesh-free method does not require a mesh to discretize the domain of the problem under consideration, and the approximate solution is constructed entirely based on a set of scattered nodes. In SMRPI technique, in contrast to those meshless methods based on weak form, there is no integration tools in this approach. Therefore, the computational complexity of SMRPI method seems to be of low order. The numerical results are in excellent agreement with exact solutions.
References
Cheng, R., Zhang, L., Liew, K.: Modeling of biological population problems using the element-free kp-ritz method. Appl. Math. Comput. 227, 274–290 (2014)
Zhang, L., Deng, Y., Liew, K.: An improved element-free galerkin method for numerical modeling of the biological population problems. Eng. Anal. Bound. Elem. 40, 181–188 (2014)
Carl, E.A.: Population control in arctic ground squirrels. Ecology. 52, 395–413 (1971)
Gurney, W., Nisbet, R.: The regulation of inhomogeneous populations. J. Theor. Biol. 52(2), 441–457 (1975)
Lu, Y.-G.: Hölder estimates of solutions of biological population equations. Appl. Math. Lett. 13(6), 123–126 (2000)
Gurtin, M.E., MacCamy, R.C.: On the diffusion of biological populations. Math. Biosci. 33(1–2), 35–49 (1977)
Aronson, D.G.: The Porous Medium Equation. In: Fasano, A., Primicerio, M. (eds.) Nonlinear Diffusion Problems, Lecture Notes in Math, vol. 1224, pp. 12–46. Springer-Verlag, New York (1986)
Jäger, W., Lu, Y.: Global regularity of solution for general degenerate parabolic equations in 1-d. J. Diff. Equ. 140(2), 365–377 (1997)
Giuggioli, L., Kenkre, V.: Analytic solutions of a nonlinear convective equation in population dynamics. Phys. D Nonlinear Phenom. 183(3), 245–259 (2003)
Shakeri, F., Dehghan, M.: Numerical solution of a biological population model using hes variational iteration method. Comput. Math. Appl. 54(7), 1197–1209 (2007)
Belytschko, T., Lu, Y.Y., Gu, L.: Element-free Galerkin methods. Int. J. Numer. Methods Eng. 37(2), 229–256 (1994)
Belytschko, T., Lu, Y.Y., Gu, L.: Element free Galerkin methods for static and dynamic fracture. Int. J. Solids Struct. 32, 2547–2570 (1995)
Duan, Y., Tan, Y.: A meshless Galerkin method for Dirichlet problems using radial basis functions. J. Comput. Appl. Math. 196(2), 394–401 (2006)
Assari, P., Adibi, H., Dehghan, M.: A meshless discrete Galerkin (MDG) method for the numerical solution of integral equations with logarithmic kernels. J. Comput. Appl. Math. 267, 160–181 (2014)
Dehghan, M., Salehi, R.: A meshless local Petrov-Galerkin method for the time-dependent Maxwell equations. J. Comput. Appl. Math. 268, 93–110 (2014)
Mirzaei, D., Dehghan, M.: Meshless local Petrov-Galerkin (MLPG) approximation to the two dimensional sine-Gordon equation. J. Comput. Appl. Math. 233(10), 2737–2754 (2010)
Fili, A., Naji, A., Duan, Y.: Coupling three-field formulation and meshless mixed Galerkin methods using radial basis functions. J. Comput. Appl. Math. 234(8), 2456–2468 (2010)
Peng, M., Li, D., Cheng, Y.: The complex variable element-free Galerkin (CVEFG) method for elasto-plasticity problems. Eng. Struct. 33(1), 127–135 (2011)
Dai, B., Cheng, Y.: An improved local boundary integral equation method for two-dimensional potential problems. Int. J. Appl. Mech. 2(2), 421–436 (2010)
Bai, F., Li, D., Wang, J., Cheng, Y.: An improved complex variable element-free Galerkin method for two-dimensional elasticity problems. Chin. Phys. B 21(2), 020204 (2012)
Atluri, S., Shen, S.: The meshless local Petrov-Galerkin (MLPG) method: a simple and less costly alternative to the finite element and boundary element methods. Comput. Model. Eng. Sci. 3, 11–51 (2002)
Kansa, E.: Multiquadrics-a scattered data approximation scheme with applications to computational fluid-dynamics. I. surface approximations and partial derivative estimates. Comput. Math. Appl. 19(8–9), 127–145 (1990)
Dehghan, M., Shokri, A.: A numerical method for solution of the two dimensional sine-Gordon equation using the radial basis functions. Math. Comput. Simul. 79, 700–715 (2008)
Jakobsson, S., Andersson, B., Edelvik, F.: Rational radial basis function interpolation with applications to antenna design. J. Comput. Appl. Math. 233(4), 889–904 (2009)
Assari, P., Adibi, H., Dehghan, M.: A meshless method for solving nonlinear two-dimensional integral equations of the second kind on non-rectangular domains using radial basis functions with error analysis. J. Comput. Appl. Math. 239, 72–92 (2013)
Zhang, Y., Tan, Y.: Meshless schemes for unsteady Navier-Stokes equations in vorticity formulation using radial basis functions. J. Comput. Appl. Math. 192(2), 328–338 (2006)
Abbasbandy, S., Ghehsareh, H.R., Hashim, I.: A meshfree method for the solution of two-dimensional cubic nonlinear schrödinger equation. Eng. Anal. Bound. Elem. 37(6), 885–898 (2013)
Abbasbandy, S., Ghehsareh, H.R., Hashim, I.: Numerical analysis of a mathematical model for capillary formation in tumor angiogenesis using a meshfree method based on the radial basis function. Eng. Anal. Bound. Elem. 36(12), 1811–1818 (2012)
Abbasbandy, S., Shirzadi, A.: A meshless method for two-dimensional diffusion equation with an integral condition. Eng. Anal. Bound. Elem. 34(12), 1031–1037 (2010)
Shirzadi, A., Sladek, V., Sladek, J.: A local integral equation formulation to solve coupled nonlinear reaction-diffusion equations by using moving least square approximation. Eng. Anal. Bound. Elem. 37, 8–14 (2013)
Dehghan, M., Ghesmati, A.: Numerical simulation of two-dimensional sine-Gordon solitons via a local weak meshless technique based on the radial point interpolation method (RPIM). Comput. Phys. Commun. 181, 772–786 (2010)
Tadeu, A., Chen, C., Antonio, J., Simoes, N.: A boundary meshless method for solving heat transfer problems using the Fourier transform. Adv. Appl. Math. Mech. 3, 572–585 (2011)
Shivanian, E.: Analysis of meshless local radial point interpolation (MLRPI) on a nonlinear partial integro-differential equation arising in population dynamics. Eng. Anal. Bound. Elem. 37, 1693–1702 (2013)
Shivanian, E.: Analysis of meshless local and spectral meshless radial point interpolation (MLRPI and SMRPI) on 3-D nonlinear wave equations. Ocean Eng. 89, 173–188 (2014)
Shivanian, E.: Meshless local Petrov-Galerkin (MLPG) method for three-dimensional nonlinear wave equations via moving least squares approximation. Eng. Anal. Bound. Elem. 50, 249–257 (2015)
Shivanian, E., Khodabandehlo, H.: Meshless local radial point interpolation (MLRPI) on the telegraph equation with purely integral conditions. Eur. Phys. J. Plus 129, 241–251 (2014)
Shivanian, E., Abbasbandy, S., Alhuthali, M.S., Alsulami, H.H.: Local integration of 2-d fractional telegraph equation via moving least squares approximation. Eng. Anal. Bound. Elem. 56, 98–105 (2015)
Hosseini, V.R., Shivanian, E., Chen, W.: Local integration of 2-D fractional telegraph equation via local radial point interpolant approximation. Eur. Phys. J. Plus 130(33), 1–21 (2015)
Aslefallah, M., Shivanian, E.: Nonlinear fractional integro-differential reaction-diffusion equation via radial basis functions. Eur. Phys. J. Plus 130(47), 1–9 (2015)
Shivanian, E.: On the convergence analysis, stability, and implementation of meshless local radial point interpolation on a class of three-dimensional wave equations. Int. J. Numer. Methods Eng. 105(2), 83–110 (2016)
Shivanian, E., Rahimi, A. Hosseini, M.: Meshless local radial point interpolation to three-dimensional wave equation with Neumann’s boundary conditions. Int. J. Comput. Math. (2015). doi:10.1080/00207160.2015.1085032
Shivanian, E.: Spectral meshless radial point interpolation (SMRPI) method to two-dimensional fractional telegraph equation. Math. Methods Appl. Sci. 39(7), 1820–1835 (2016)
Shivanian, E.: A new spectral meshless radial point interpolation (SMRPI) method: A well-behaved alternative to the meshless weak forms. Eng. Anal. Bound. Elem. 54, 1–12 (2015)
Fatahi, H., Saberi-Nadjafi, J., Shivanian, E.: A new spectral meshless radial point interpolation (smrpi) method for the two-dimensional fredholm integral equations on general domains with error analysis. J. Comput. Appl. Math. 294, 196–209 (2016)
Acknowledgments
The authors acknowledge financial support from the Imam Khomeini International University project IKIU-11657. The authors would like to thank anonymous referees for valuable suggestions.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Abbasbandy, S., Shivanian, E. Numerical simulation based on meshless technique to study the biological population model. Math Sci 10, 123–130 (2016). https://doi.org/10.1007/s40096-016-0186-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40096-016-0186-9