Abstract
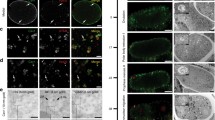
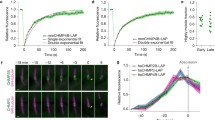
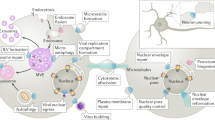
Proteins of the ESCRT (endosomal sorting complex required for transport) complex function in membrane fission processes, such as multivesicular body (MVBs) formation, the terminal stages of cytokinesis, and separation of enveloped viruses from the plasma membrane. In mammalian cells, the machinery consists of a network of more than 20 proteins, organized into three complexes (ESCRT-I, -II, and -III), and other associated proteins such as the ATPase vacuolar protein sorting 4 (Vps4). Early biochemical studies of MVBs biogenesis in yeast support a model of sequential recruitment of ESCRT complexes on membranes. Live-cell imaging of ESCRT protein dynamics during viral budding and cytokinesis now reveal that this long-standing model of sequential assembly and disassembly holds true in mammalian cells.





Similar content being viewed by others
References
Carlton JG, Martin-Serrano J (2007) Parallels between cytokinesis and retroviral budding: a role for the ESCRT machinery. Science 316(5833):1908–1912
Morita E et al (2007) Human ESCRT and ALIX proteins interact with proteins of the midbody and function in cytokinesis. EMBO J 26(19):4215–4227
Morita E et al (2010) Human ESCRT-III and VPS4 proteins are required for centrosome and spindle maintenance. Proc Natl Acad Sci USA 107(29):12889–12894
Katzmann DJ, Babst M, Emr SD (2001) Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106(2):145–155
Caballe A, Martin-Serrano J (2011) ESCRT machinery and cytokinesis: the road to daughter cell separation. Traffic 12(10):1318–1326
Hurley JH, Hanson PI (2010) Membrane budding and scission by the ESCRT machinery: it’s all in the neck. Nat Rev Mol Cell Biol 11(8):556–566
Henne WM, Buchkovich NJ, Emr SD (2011) The ESCRT pathway. Dev Cell 21(1):77–91
Guizetti J, Gerlich DW (2012) ESCRT-III polymers in membrane neck constriction. Trends Cell Biol 22(3):133–140
Elia N et al (2011) Dynamics of endosomal sorting complex required for transport (ESCRT) machinery during cytokinesis and its role in abscission. Proc Natl Acad Sci USA 108(12):4846–4851
Guizetti J et al (2011) Cortical constriction during abscission involves helices of ESCRT-III-dependent filaments. Science 331(6024):1616–1620
Jouvenet N et al (2011) Dynamics of ESCRT protein recruitment during retroviral assembly. Nat Cell Biol 13(4):394–401
Baumgartel V et al (2011) Live-cell visualization of dynamics of HIV budding site interactions with an ESCRT component. Nat Cell Biol 13(4):469–474
Babst M et al (2000) Mammalian tumor susceptibility gene 101 (TSG101) and the yeast homologue, Vps23p, both function in late endosomal trafficking. Traffic 1(3):248–258
Bishop N, Woodman P (2001) TSG101/mammalian VPS23 and mammalian VPS28 interact directly and are recruited to VPS4-induced endosomes. J Biol Chem 276(15):11735–11742
Eastman SW et al (2005) Identification of human VPS37C, a component of endosomal sorting complex required for transport-I important for viral budding. J Biol Chem 280(1):628–636
Morita E et al (2007) Identification of human MVB12 proteins as ESCRT-I subunits that function in HIV budding. Cell Host Microbe 2(1):41–53
Stefani F et al (2011) UBAP1 is a component of an endosome-specific ESCRT-I complex that is essential for MVB sorting. Curr Biol 21(14):1245–1250
Stuchell MD et al (2004) The human endosomal sorting complex required for transport (ESCRT-I) and its role in HIV-1 budding. J Biol Chem 279(34):36059–36071
Agromayor M et al (2012) The UBAP1 Subunit of ESCRT-I Interacts with ubiquitin via a SOUBA domain. Structure 20:414–418
Kim J et al (2005) Structural basis for endosomal targeting by the Bro1 domain. Dev Cell 8(6):937–947
Lee S et al (2007) Structural basis for viral late-domain binding to Alix. Nat Struct Mol Biol 14(3):194–199
Fisher RD et al (2007) Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell 128(5):841–852
McMahon HT, Gallop JL (2005) Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature 438(7068):590–596
Odorizzi G et al (2003) Bro1 is an endosome-associated protein that functions in the MVB pathway in Saccharomyces cerevisiae. J Cell Sci 116(Pt 10):1893–1903
McCullough J et al (2008) ALIX–CHMP4 interactions in the human ESCRT pathway. Proc Natl Acad Sci USA 105(22):7687–7691
Martin-Serrano J, Zang T, Bieniasz PD (2003) Role of ESCRT-I in retroviral budding. J Virol 77(8):4794–4804
Martin-Serrano J et al (2003) Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc Natl Acad Sci USA 100(21):12414–12419
Lee HH et al (2008) Midbody targeting of the ESCRT machinery by a noncanonical coiled coil in CEP55. Science 322(5901):576–580
Langelier C et al (2006) Human ESCRT-II complex and its role in human immunodeficiency virus type 1 release. J Virol 80(19):9465–9480
Wollert T et al (2009) Membrane scission by the ESCRT-III complex. Nature 458(7235):172–177
Shim S, Kimpler LA, Hanson PI (2007) Structure/function analysis of four core ESCRT-III proteins reveals common regulatory role for extreme C-terminal domain. Traffic 8(8):1068–1079
Zamborlini A et al (2006) Release of autoinhibition converts ESCRT-III components into potent inhibitors of HIV-1 budding. Proc Natl Acad Sci USA 103(50):19140–19145
Lata S et al (2008) Helical structures of ESCRT-III are disassembled by VPS4. Science 321(5894):1354–1357
Peel S et al (2010) Divergent pathways lead to ESCRT-III-catalyzed membrane fission. Trends Biochem Sci 36(4):199–210
Bodon G et al (2012) Charged multivesicular body protein 2B (CHMP2B) of the endosomal sorting complex required for transport-III (ESCRT-III) polymerizes into helical structures deforming the plasma membrane. J Biol Chem 286(46):40276–40286
Fabrikant G et al (2009) Computational model of membrane fission catalyzed by ESCRT-III. PLoS Comput Biol 5(11):e1000575
Agromayor M et al (2009) Essential role of hIST1 in cytokinesis. Mol Biol Cell 20(5):1374–1387
Bajorek M et al (2009) Biochemical analyses of human IST1 and its function in cytokinesis. Mol Biol Cell 20(5):1360–1373
Obita T et al (2007) Structural basis for selective recognition of ESCRT-III by the AAA ATPase Vps4. Nature 449(7163):735–739
Stuchell-Brereton MD et al (2007) ESCRT-III recognition by VPS4 ATPases. Nature 449(7163):740–744
Scott A et al (2005) Structural and mechanistic studies of VPS4 proteins. EMBO J 24(20):3658–3669
Gonciarz MD et al (2008) Biochemical and structural studies of yeast Vps4 oligomerization. J Mol Biol 384(4):878–895
Yu Z et al (2008) Cryo-EM structure of dodecameric Vps4p and its 2:1 complex with Vta1p. J Mol Biol 377(2):364–377
Hartmann C et al (2008) Vacuolar protein sorting: two different functional states of the AAA-ATPase Vps4p. J Mol Biol 377(2):352–363
Landsberg MJ et al (2009) Three-dimensional structure of AAA ATPase Vps4: advancing structural insights into the mechanisms of endosomal sorting and enveloped virus budding. Structure 17(3):427–437
Inoue M et al (2008) Nucleotide-dependent conformational changes and assembly of the AAA ATPase SKD1/VPS4B. Traffic 9(12):2180–2189
Babst M et al (2002) Escrt-III: an endosome-associated hetero oligomeric protein complex required for mvb sorting. Dev Cell 3(2):271–282
Babst M et al (2002) Endosome-associated complex, ESCRT-II, recruits transport machinery for protein sorting at the multivesicular body. Dev Cell 3(2):283–289
Wollert T, Hurley JH (2010) Molecular mechanism of multivesicular body biogenesis by ESCRT complexes. Nature 464(7290):864–869
Saksena S et al (2009) Functional reconstitution of ESCRT-III assembly and disassembly. Cell 136(1):97–109
Ghazi-Tabatabai S et al (2008) Structure and disassembly of filaments formed by the ESCRT-III subunit Vps24. Structure 16(9):1345–1356
Shim S, Merrill SA, Hanson PI (2008) Novel interactions of ESCRT-III with LIP5 and VPS4 and their implications for ESCRT-III disassembly. Mol Biol Cell 19(6):2661–2672
Azmi I et al (2006) Recycling of ESCRTs by the AAA-ATPase Vps4 is regulated by a conserved VSL region in Vta1. J Cell Biol 172(5):705–717
Samson RY et al (2008) A role for the ESCRT system in cell division in Archaea. Science 322(5908):1710–1713
Samson RY et al (2011) Molecular and structural basis of ESCRT-III recruitment to membranes during archaeal cell division. Mol Cell 41(2):186–196
Moriscot C et al (2011) Crenarchaeal CdvA forms double-helical filaments containing DNA and interacts with ESCRT-III-like CdvB. PLoS ONE 6(7):e21921
Felder S et al (1990) Kinase activity controls the sorting of the epidermal growth factor receptor within the multivesicular body. Cell 61(4):623–634
Futter CE et al (1996) Multivesicular endosomes containing internalized EGF–EGF receptor complexes mature and then fuse directly with lysosomes. J Cell Biol 132(6):1011–1023
Teis D, Saksena S, Emr SD (2008) Ordered assembly of the ESCRT-III complex on endosomes is required to sequester cargo during MVB formation. Dev Cell 15(4):578–589
Strack B et al (2003) AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114(6):689–699
Glotzer M (2009) The 3Ms of central spindle assembly: microtubules, motors and MAPs. Nat Rev Mol Cell Biol 10(1):9–20
Skop AR et al (2004) Dissection of the mammalian midbody proteome reveals conserved cytokinesis mechanisms. Science 305(5680):61–66
Fabbro M et al (2005) Cdk1/Erk2- and Plk1-dependent phosphorylation of a centrosome protein, Cep55, is required for its recruitment to midbody and cytokinesis. Dev Cell 9(4):477–488
Martinez-Garay I et al (2006) The novel centrosomal associated protein CEP55 is present in the spindle midzone and the midbody. Genomics 87(2):243–253
Zhao WM, Seki A, Fang G (2006) Cep55, a microtubule-bundling protein, associates with centralspindlin to control the midbody integrity and cell abscission during cytokinesis. Mol Biol Cell 17(9):3881–3896
Guizetti J et al (2010) Correlative time-lapse imaging and electron microscopy to study abscission in HeLa cells. Methods Cell Biol 96:591–601
Steigemann P et al (2009) Aurora B-mediated abscission checkpoint protects against tetraploidization. Cell 136(3):473–484
Jin Y et al (2005) The fission yeast homolog of the human transcription factor EAP30 blocks meiotic spindle pole body amplification. Dev Cell 9(1):63–73
Carlton JG, Agromayor M, Martin-Serrano J (2008) Differential requirements for Alix and ESCRT-III in cytokinesis and HIV-1 release. Proc Natl Acad Sci USA 105(30):10541–10546
Carlton JG et al (2012) ESCRT-III governs the Aurora B-mediated abscission checkpoint through CHMP4C. Science 336:220–225
Hanson PI et al (2008) Plasma membrane deformation by circular arrays of ESCRT-III protein filaments. J Cell Biol 180(2):389–402
Pires R et al (2009) A crescent-shaped ALIX dimer targets ESCRT-III CHMP4 filaments. Structure 17(6):843–856
Briggs JA et al (2004) The stoichiometry of Gag protein in HIV-1. Nat Struct Mol Biol 11(7):672–675
Jacks T et al (1988) Characterization of ribosomal frame shifting in HIV-1 gag-pol expression. Nature 331(6153):280–283
Zhu P et al (2003) Electron tomography analysis of envelope glycoprotein trimers on HIV and simian immunodeficiency virus virions. Proc Natl Acad Sci USA 100(26):15812–15817
Chen J et al (2009) High efficiency of HIV-1 genomic RNA packaging and heterozygote formation revealed by single virion analysis. Proc Natl Acad Sci USA 106(32):13535–13540
Larson DR et al (2005) Visualization of retrovirus budding with correlated light and electron microscopy. Proc Natl Acad Sci USA 102(43):15453–15458
Goff SP (2001) Retroviridae: the viruses and their replication. In: Knipe DM, Howley PM (eds) Fields virology. Lippincott Williams and Wilkins, Philadelphia, pp 1999–2070
Kutluay SB, Bieniasz PD (2010) Analysis of the initiating events in HIV-1 particle assembly and genome packaging. PLoS Pathog 6(11):e1001200
Jouvenet N et al (2011) Cell biology of retroviral RNA packaging. RNA Biol 8(4):572–580
Jouvenet N, Simon SM, Bieniasz PD (2009) Imaging the interaction of HIV-1 genomes and Gag during assembly of individual viral particles. Proc Natl Acad Sci USA 106(45):19114–19119
Kemler I, Meehan A, Poeschla EM (2010) Live-cell coimaging of the genomic RNAs and Gag proteins of two lentiviruses. J Virol 84(13):6352–6366
Jouvenet N et al (2006) Plasma membrane is the site of productive HIV-1 particle assembly. PLoS Biol 4(12):e435
Finzi A et al (2007) Productive human immunodeficiency virus type 1 assembly takes place at the plasma membrane. J Virol 81(14):7476–7490
Welsch S et al (2007) HIV-1 buds predominantly at the plasma membrane of primary human macrophages. PLoS Pathog 3(3):e36
Chen BJ, Lamb RA (2008) Mechanisms for enveloped virus budding: can some viruses do without an ESCRT? Virology 372(2):221–232
Bieniasz PD (2009) The cell biology of HIV-1 virion genesis. Cell Host Microbe 5(6):550–558
Carlton JG, Martin-Serrano J (2009) The ESCRT machinery: new functions in viral and cellular biology. Biochem Soc Trans 37(Pt 1):195–199
Martin-Serrano J, Zang T, Bieniasz PD (2001) HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat Med 7(12):1313–1319
Garrus JE et al (2001) Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107(1):55–65
von Schwedler UK et al (2003) The protein network of HIV budding. Cell 114(6):701–713
Martin-Serrano J et al (2005) HECT ubiquitin ligases link viral and cellular PPXY motifs to the vacuolar protein-sorting pathway. J Cell Biol 168(1):89–101
Zhadina M, Bieniasz PD (2010) Functional interchangeability of late domains, late domain cofactors and ubiquitin in viral budding. PLoS Pathog 6(10):e1001153
Weiss ER et al (2010) Rescue of HIV-1 release by targeting widely divergent NEDD4-type ubiquitin ligases and isolated catalytic HECT domains to Gag. PLoS Pathog 6(9):e1001107
Rauch S, Martin-Serrano J (2011) Multiple interactions between the ESCRT machinery and arrestin-related proteins: implications for PPXY-dependent budding. J Virol 85(7):3546–3556
VerPlank L et al (2001) Tsg101, a homologue of ubiquitin-conjugating (E2) enzymes, binds the L domain in HIV type 1 Pr55(Gag). Proc Natl Acad Sci USA 98(14):7724–7729
Popov S et al (2009) Divergent Bro1 domains share the capacity to bind human immunodeficiency virus type 1 nucleocapsid and to enhance virus-like particle production. J Virol 83(14):7185–7193
Popova E, Popov S, Gottlinger HG (2010) Human immunodeficiency virus type 1 nucleocapsid p1 confers ESCRT pathway dependence. J Virol 84(13):6590–6597
Dussupt V et al (2009) The nucleocapsid region of HIV-1 Gag cooperates with the PTAP and LYPXnL late domains to recruit the cellular machinery necessary for viral budding. PLoS Pathog 5(3):e1000339
Fujii K et al (2009) Functional role of Alix in HIV-1 replication. Virology 391(2):284–292
Morita E et al (2011) ESCRT-III protein requirements for HIV-1 budding. Cell Host Microbe 9(3):235–242
Usami Y, Popov S, Gottlinger HG (2007) Potent rescue of human immunodeficiency virus type 1 late domain mutants by ALIX/AIP1 depends on its CHMP4 binding site. J Virol 81(12):6614–6622
Dussupt V et al (2010) Basic residues in the nucleocapsid domain of Gag are critical for late events of HIV-1 budding. J Virol 85(5):2304–2315
Leroux C, Cadore JL, Montelaro RC (2004) Equine infectious anemia virus (EIAV): what has HIV’s country cousin got to tell us? Vet Res 35(4):485–512
Simon SM (2009) Partial internal reflections on total internal reflection fluorescent microscopy. Trends Cell Biol 19(11):661–668
Jouvenet N, Bieniasz PD, Simon SM (2008) Imaging the biogenesis of individual HIV-1 virions in live cells. Nature 454(7201):236–240
Ivanchenko S et al (2009) Dynamics of HIV-1 assembly and release. PLoS Pathog 5(11):e1000652
Fusco D et al (2003) Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Curr Biol 13(2):161–167
Lin Y et al (2005) Interaction of the mammalian endosomal sorting complex required for transport (ESCRT) III protein hSnf7-1 with itself, membranes, and the AAA+ ATPase SKD1. J Biol Chem 280(13):12799–12809
Howard TL et al (2001) CHMP1 functions as a member of a newly defined family of vesicle trafficking proteins. J Cell Sci 114(Pt 13):2395–2404
Miesenbock G, De Angelis DA, Rothman JE (1998) Visualizing secretion and synaptic transmission with pH-sensitive green fluorescent proteins. Nature 394(6689):192–195
Acknowledgments
I thank the ‘Centre National de la Recherche Scientifique’ for support, Sandy Simon for critical reading of the manuscript, and Juan Martin-Serrano for comments and access to unpublished data.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jouvenet, N. Dynamics of ESCRT proteins. Cell. Mol. Life Sci. 69, 4121–4133 (2012). https://doi.org/10.1007/s00018-012-1035-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-012-1035-0