Abstract
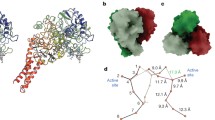
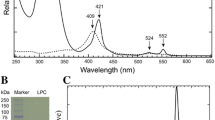
SoxAX cytochromes are heme-thiolate proteins that play a key role in bacterial thiosulfate oxidation, where they initiate the reaction cycle of a multi-enzyme complex by catalyzing the attachment of sulfur substrates such as thiosulfate to a conserved cysteine present in a carrier protein. SoxAX proteins have a wide phylogenetic distribution and form a family with at least three distinct types of SoxAX protein. The types of SoxAX cytochromes differ in terms of the number of heme groups present in the proteins (there are diheme and triheme versions) as well as in their subunit structure. While two of the SoxAX protein types are heterodimers, the third group contains an additional subunit, SoxK, that stabilizes the complex of the SoxA and SoxX proteins. Crystal structures are available for representatives of the two heterodimeric SoxAX protein types and both of these have shown that the cysteine ligand to the SoxA active site heme carries a modification to a cysteine persulfide that implicates this ligand in catalysis. EPR studies of SoxAX proteins have also revealed a high complexity of heme dependent signals associated with this active site heme; however, the exact mechanism of catalysis is still unclear at present, as is the exact number and types of redox centres involved in the reaction.







Similar content being viewed by others
References
Bamford VA, Bruno S, Rasmussen T, Appia-Ayme C, Cheesman MR, Berks BC, Hemmings AM (2002) Structural basis for the oxidation of thiosulfate by a sulfur cycle enzyme. EMBO J 21(21):5599–5610
Lu WP, Kelly DP (1988) Respiration-driven proton translocation in Thiobacillus versutus and the role of the periplasmic thiosulphate-oxidizing enzyme system. Arch Microbiol 149:297–302
Omura T (2005) Heme-thiolate proteins. Biochem Biophys Res Comm 338(1):404–409
Green MT (2009) CH bond activation in heme proteins: the role of thiolate ligation in cytochrome P450. Curr Opin Chem Biol 13(1):84–88
Pazicni S, Cherney MM, Lukat-Rodgers GS, Oliveriusova J, Rodgers KR, Kraus JP, Burstyn JN (2005) The heme of cystathionine beta-synthase likely undergoes a thermally induced redox-mediated ligand switch. Biochemistry 44(51):16785–16795
Youn H, Conrad M, Chung SY, Roberts GP (2006) Roles of the heme and heme ligands in the activation of CooA, the CO-sensing transcriptional activator. Biochem Biophys Res Comm 348(2):345–350
Dhawan IK, Shelver D, Thorsteinsson MV, Roberts GP, Johnson MK (1999) Probing the heme axial ligation in the CO-sensing CooA protein with magnetic circular dichroism spectroscopy. Biochemistry 38(39):12805–12813
Alric J, Tsukatani Y, Yoshida M, Matsuura K, Shimada K, Hienerwadel R, Schoepp-Cothenet B, Nitschke W, Nagashima KVP, Vermeglio A (2004) Structural and functional characterization of the unusual triheme cytochrome bound to the reaction center of Rhodovulum sulfidophilum. J Biol Chem 279(25):26090–26097
Grein F, Venceslau SS, Schneider L, Hildebrandt P, Todorovic S, Pereira IAC, Dahl C (2010) DsrJ, an essential part of the DsrMKJOP transmembrane complex in the purple sulfur bacterium Allochromatium vinosum, is an unusual triheme cytochrome c. Biochemistry 49(38):8290–8299
Grein F, Pereira IAC, Dahl C (2010) Biochemical characterization of individual components of the Allochromatium vinosum DsrMKJOP transmembrane complex aids understanding of complex function in vivo. J Bacteriol 192(24):6369–6377
Lu WP, Swoboda BEP, Kelly DP (1985) Properties of the thiosulphate oxidising multi-enzyme system from Thiobacillus versutus. Biochim Biophys Acta 828:116–122
Lu WP, Kelly DP (1984) Properties and role of sulphite:cytochrome c oxidoreductase purified from Thiobacillus versutus. J Gen Microbiol 130:1683–1692
Lu W-P, Kelly DP (1984) Purification and characterization of two essential cytochromes of the thiosulphate-oxidizing multi-enzyme system from Thiobacillus A2 (Thiobacillus versutus). Biochim Biophys Acta 765(2):106–117
Katayama Y, Hiraishi A, Kuraishi H (1995) Paracoccus thiocyanatus sp. nov., a new species of thiocyanate- utilizing facultative chemolithotroph, and transfer of Thiobacillus versutus to the genus Paracoccus as Paracoccus versutus comb. nov. with emendation of the genus. Microbiology 141:1469–1477
Kelly DP, Shergill JK, Lu WP, Wood AP (1997) Oxidative metabolism of inorganic sulfur compounds by bacteria. Antonie Van Leeuwenhoek 71(1–2):95–107
Mittenhuber G, Sonomoto K, Egert M, Friedrich CG (1991) Identification of the DNA region responsible for sulfur-oxidizing ability of Thiosphaera pantotropha. J Bacteriol 173(2):7340–7344
Wodara C, Kostka S, Egert M, Kelly DP, Friedrich CG (1994) Identification and sequence analysis of the soxB gene essential for sulfur oxidation of Paracoccus denitrificans GB17. J Bacteriol 176:6188–6191
Ludwig W, Mittenhuber G, Friedrich CG (1993) Transfer of Thiosphaera pantotropha to Paracoccus denitrificans. Int J Sys Bacteriol 43(2):363–367
Quentmeier A, Li L, Friedrich CG (2008) Identification of two inactive forms of the central sulfur cycle protein SoxYZ of Paracoccus pantotrophus. FEBS Lett 582(25–26):3701–3704
Reijerse EJ, Sommerhalter M, Hellwig P, Quentmeier A, Rother D, Laurich C, Bothe E, Lubitz W, Friedrich CG (2007) The unusual redox properties of SoxXA, a novel c-type heme-enzyme essential for chemotrophic sulfur-oxidation of Paracoccus pantotrophus. Biochemistry 46(26):7804–7810
Bardischewsky F, Quentmeier A, Friedrich CG (2006) The flavoprotein SoxF functions in chemotrophic thiosulfate oxidation of Paracoccus pantotrophus in vivo and in vitro. FEMS Microbiol Lett 258(1):121–126
Bardischewsky F, Quentmeier A, Rother D, Hellwig P, Kostka S, Friedrich CG (2005) Sulfur dehydrogenase of Paracoccus pantotrophus : the heme-2 domain of the molybdoprotein cytochrome c complex is dispensable for catalytic activity. Biochemistry 44(18):7024–7034
Friedrich CG, Rother D, Bardischewsky F, Quentmeier A, Fischer J (2001) Oxidation of reduced inorganic sulfur compounds by bacteria: emergence of a common mechanisms? Appl Environ Microbiol 67(7):2873–2882
Quentmeier A, Kraft R, Kostka S, Klockenkamper R, Friedrich CG (2000) Characterization of a new type of sulfite dehydrogenase from Paracoccus pantotrophus GB17. Arch Microbiol 173(2):117–125
Rother D, Ringk J, Friedrich CG (2008) Sulfur oxidation of Paracoccus pantotrophus: the sulfur-binding protein SoxYZ is the target of the periplasmic thiol-disulfide oxidoreductase SoxS. Microbiology 154:1980–1988
Bardischewsky F, Fischer J, Holler B, Friedrich CG (2006) SoxV transfers electrons to the periplasm of Paracoccus pantotrophus—an essential reaction for chemotrophic sulfur oxidation. Microbiology 152:465–472
Rother D, Orawski G, Bardischewsky F, Friedrich CG (2005) SoxRS-mediated regulation of chemotrophic sulfur oxidation in Paracoccus pantotrophus. Microbiology 151:1707–1716
Bardischewsky F, Friedrich CG (2001) Identification of ccdA in Paracoccus pantotrophus GB17: disruption of ccdA causes complete deficiency in c-type cytochromes. J Bacteriol 183(1):257–263
Bardischewsky F, Friedrich CG (2001) The shxVW locus is essential for oxidation of inorganic sulfur and molecular hydrogen by Paracoccus pantotrophus GB17: a novel function for lithotrophy. FEMS Microbiol Lett 202:215–220
Friedrich CG, Bardischewsky F, Rother D, Quentmeier A, Fischer J (2005) Prokaryotic sulfur oxidation. Curr Opin Microbiol 8(3):253–259
Verte F, Kostanjevecki V, De Smet L, Meyer TE, Cusanovich MA, Van Beeumen JJ (2002) Identification of a thiosulfate utilization gene cluster from the green phototrophic bacterium Chlorobium limicola. Biochemistry 41(9):2932–2945
Klarskov K, Verte F, VanDriessche G, Meyer TE, Cusanovich MA, Van Beeumen J (1998) The primary structure of soluble cytochrome c 551 from the photographic green sulfur bacterium Chlorobium limicola, strain Tassajara, reveals a novel c-type cytochrome. Biochemistry 37(30):10555–10562
Frigaard NU, Dahl C, Robert KP (2008) Sulfur metabolism in phototrophic sulfur bacteria. In: Advances in Microbial Physiology, vol 54. Academic Press, pp 103–200
Hensen D, Sperling D, Truper HG, Brune DC, Dahl C (2006) Thiosulphate oxidation in the phototrophic sulphur bacterium Allochromatium vinosum. Mol Microbiol 62(3):794–810
Sauve V, Bruno S, Berks BC, Hemmings AM (2007) The SoxYZ complex carries sulfur cycle intermediates on a peptide swinging arm. J Biol Chem 282(32):23194–23204
Ghosh W, Roy P (2007) Chemolithoautotrophic oxidation of thiosulfate, tetrathionate and thiocyanate by a novel rhizobacterium belonging to the genus Paracoccus. FEMS Microbiol Lett 270(1):124–131
Sauve V, Roversi P, Leath KJ, Garman EF, Antrobus R, Lea SM, Berks BC (2009) Mechanism for the hydrolysis of a sulfur–sulfur bond based on the crystal structure of the thiosulfohydrolase SoxB. J Biol Chem 284(32):21707–21718
Zander U, Faust A, Klink BU, de Sanctis D, Panjikar S, Quentmeier A, Bardischewsky F, Friedrich CG, Scheidig AJ (2011) Structural basis for the oxidation of protein-bound sulfur by the sulfur cycle molybdohemo-enzyme sulfane dehydrogenase SoxCD. J Biol Chem 286(10):8349–8360. doi:10.1074/jbc.M110.193631
Frigaard NU, Dahl C (2009) Sulfur metabolism in phototrophic sulfur bacteria. Adv Microb Phys 54:103–200
Kappler U, Aguey-Zinsou KF, Hanson GR, Bernhardt PV, McEwan AG (2004) Cytochrome c551 from Starkeya novella: characterization, spectroscopic properties, and phylogeny of a diheme protein of the SoxAX family. J Biol Chem 279(8):6252–6260
Ogawa T, Furusawa T, Nomura R, Seo D, Hosoya-Matsuda N, Sakurai H, Inoue K (2008) SoxAX binding protein, a novel component of the thiosulfate-oxidizing multienzyme system in the green sulfur bacterium Chlorobium tepidum. J Bacteriol 190(18):6097–6110
Orawski G, Bardischewsky F, Quentmeier A, Rother D, Friedrich CG (2007) The periplasmic thioredoxin SoxS plays a key role in activation in vivo of chemotrophic sulfur oxidation of Paracoccus pantotrophus. Microbiology 153:1081–1086
Gregersen LH, Bryant DA, Frigaard N-U (2011) Mechanisms and evolution of oxidative sulfur metabolism in green sulfur bacteria. Frontiers Microbiol 2:116. doi:10.3389/fmicb.2011.00116
Sakurai H, Ogawa T, Shiga M, Inoue K (2010) Inorganic sulfur oxidizing system in green sulfur bacteria. Photosynth Res 104(2–3):163–176. doi:10.1007/s11120-010-9531-2
Kilmartin JR, Maher MJ, Krusong K, Noble CJ, Hanson GR, Bernhardt PV, Riley MJ, Kappler U (2011) Insights into structure and function of the active site of SoxAX cytochromes. J Biol Chem 286(28):24872–24881
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739
Rother D, Henrich HJ, Quentmeier A, Bardischewsky F, Friedrich CG (2001) Novel genes of the sox gene cluster, mutagenesis of the flavoprotein SoxF, and evidence for a general sulfur-oxidizing system in Paracoccus pantotrophus GB17. J Bacteriol 183(15):4499–4508
Dambe T, Quentmeier A, Rother D, Friedrich C, Scheidig AJ (2005) Structure of the cytochrome complex SoxXA of Paracoccus pantotrophus, a heme enzyme initiating chemotrophic sulfur oxidation. J Struct Biol 152(3):229–234
Friedrich CG, Quentmeier A, Bardischewsky F, Rother D, Kraft R, Kostka S, Prinz H (2000) Novel genes coding for lithotrophic sulfur oxidation of Paracoccus pantotrophus GB17. J Bacteriol 182(17):4677–4687
Kappler U, Bernhardt PV, Kilmartin J, Riley MJ, Teschner J, McKenzie KJ, Hanson GR (2008) SoxAX cytochromes, a new type of heme copper protein involved in bacterial energy generation from sulfur compounds. J Biol Chem 283(32):22206–22214
Frausto da Silva JJR, Williams RJP (2001) The biological chemistry of the elements—the inorganic chemistry of life. Oxford University Press, Oxford
Cheesman MR, Little PJ, Berks BC (2001) Novel heme ligation in a c-type cytochrome involved in thiosulfate oxidation: EPR and MCD of SoxAX from Rhodovulum sulfidophilum. Biochemistry 40:10562–10569
Kappler U, Hanson GR, Jones A, McEwan AG (2005) A recombinant diheme SoxAX cytochrome-implications for the relationship between EPR signals and modified heme-ligands. FEBS Lett 579:2491–2498
Welte C, Hafner S, Kratzer C, Quentmeier A, Friedrich CG, Dahl C (2009) Interaction between Sox proteins of two physiologically distinct bacteria and a new protein involved in thiosulfate oxidation. FEBS Lett 583(8):1281–1286
Multhaup G, Schlicksupp A, Hesse L, Beher D, Ruppert T, Masters CL, Beyreuther K (1996) The amyloid precursor protein of Alzheimer’s disease in the reduction of copper(II) to copper(I). Science 271(5254):1406–1409
Acknowledgments
This work was supported by a fellowship (Australian Research Fellowship, DP0878525) from the Australian Research Council to UK. MJM is supported by a La Trobe Institute for Molecular Science Senior Research Fellowship.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
18_2012_1098_MOESM1_ESM.pdf
Figure S1 Phylogenetic relationship of SoxA proteins. Two hundred sixteen SoxA amino acid sequences were analyzed using Mega 5.0. The tree shown was generated using the Neighbor-joining algorithm (Poisson model; uniform rate of evolution for all sites; gap treatment: pairwise-deletion; robustness testing: bootstrap method with 500 resampling cycles). This figure shows a tree that is identical to that shown in Fig. 3 but all branches are fully expanded. (PDF 58 kb)
Rights and permissions
About this article
Cite this article
Kappler, U., Maher, M.J. The bacterial SoxAX cytochromes. Cell. Mol. Life Sci. 70, 977–992 (2013). https://doi.org/10.1007/s00018-012-1098-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-012-1098-y