Abstract
Background
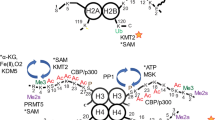
Methyl-dependent regulation of transcription has expanded from a traditional focus on histones to encompass transcription factor modulation. While the Set7 lysine methyltransferase is associated with pro-inflammatory gene expression in vascular endothelial cells, genome-wide regulatory roles remain to be investigated. From initial characterization of Set7 as specific for methyl-lysine 4 of H3 histones (H3K4m1), biochemical activity toward non-histone substrates has revealed additional mechanisms of gene regulation.
Results
mRNA-Seq revealed transcriptional deregulation of over 8,000 genes in an endothelial model of Set7 knockdown. Gene ontology identified up-regulated pathways involved in developmental processes and extracellular matrix remodeling, whereas pathways regulating the inflammatory response as well as nitric oxide signaling were down-regulated. Chromatin maps derived from ChIP-Seq profiling of H3K4m1 identified several hundred loci with loss of H3K4m1 at gene regulatory elements associated with an unexpectedly subtle effect on gene expression. Transcription factor network analysis implicated six previously described Set7 substrates in mRNA-Seq changes, and we predict that Set7 post-translationally regulates other transcription factors associated with vascular endothelial gene expression through the presence of Set7 amino acid methylation motifs.
Conclusion
We describe a role for Set7 in regulating developmental pathways and response to stimuli (inflammation/immune response) in human endothelial cells of vascular origin. Set7-dependent gene expression changes that occurred independent of H3K4m1 may involve transcription factor lysine methylation events. The method of mapping measured transcriptional changes to transcription factors to identify putative substrates with strong associations to functional changes is applicable to substrate prediction for other broad-substrate histone modifiers.









Similar content being viewed by others
References
Schwartz D, Chou MF, Church GM (2009) Predicting protein post-translational modifications using meta-analysis of proteome scale data sets. Mol Cell Proteomics 8(2):365–379
Basu A et al (2009) Proteome-wide prediction of acetylation substrates. Proc Natl Acad Sci USA 106(33):13785–13790
Dhayalan A et al (2011) Specificity analysis-based identification of new methylation targets of the SET7/9 protein lysine methyltransferase. Chem Biol 18(1):111–120
Rathert P et al (2008) Protein lysine methyltransferase G9a acts on non-histone targets. Nat Chem Biol 4(6):344–346
Rafehi H et al (2014) Vascular histone deacetylation by pharmacological HDAC inhibition. Genome Res
Wang H et al (2001) Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol Cell 8(6):1207–1217
Martens JH et al (2003) Cascade of distinct histone modifications during collagenase gene activation. Mol Cell Biol 23(5):1808–1816
Brasacchio D et al (2009) Hyperglycemia induces a dynamic cooperativity of histone methylase and demethylase enzymes associated with gene-activating epigenetic marks that coexist on the lysine tail. Diabetes 58(5):1229–1236
El-Osta A et al (2008) Transient high glucose causes persistent epigenetic changes and altered gene expression during subsequent normoglycemia. J Exp Med 205(10):2409–2417
Okabe J et al (2012) Distinguishing hyperglycemic changes by Set7 in vascular endothelial cells. Circ Res 110(8):1067–1076
Heintzman ND et al (2009) Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459(7243):108–112
Koch CM et al (2007) The landscape of histone modifications across 1% of the human genome in five human cell lines. Genome Res 17(6):691–707
Heintzman ND et al (2007) Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39(3):311–318
Nishioka K et al (2002) Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes Dev 16(4):479–489
Wang D et al (2013) Methylation of SUV39H1 by SET7/9 results in heterochromatin relaxation and genome instability. Proc Natl Acad Sci USA 110(14):5516–5521
Chuikov S et al (2004) Regulation of p53 activity through lysine methylation. Nature 432(7015):353–360
Ivanov GS et al (2007) Methylation-acetylation interplay activates p53 in response to DNA damage. Mol Cell Biol 27(19):6756–6769
Kurash JK et al (2008) Methylation of p53 by Set7/9 mediates p53 acetylation and activity in vivo. Mol Cell 29(3):392–400
Ea CK, Baltimore D (2009) Regulation of NF-kappaB activity through lysine monomethylation of p65. Proc Natl Acad Sci USA 106(45):18972–18977
Yang XD et al (2009) Negative regulation of NF-kappaB action by Set9-mediated lysine methylation of the RelA subunit. EMBO J 28(8):1055–1066
Calnan DR et al (2012) Methylation by Set9 modulates FoxO3 stability and transcriptional activity. Aging (Albany NY) 4(7):462–479
Keating S, El-Osta A (2013) Transcriptional regulation by the Set7 lysine methyltransferase. Epigenetics 8(4)
Subramanian A et al (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102(43):15545–15550
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25(14):1754–1760
Quinlan AR, Hall IM (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26(6):841–842
Zhang Y et al (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol 9(9):R137
Yang J et al (2010) Reversible methylation of promoter-bound STAT3 by histone-modifying enzymes. Proc Natl Acad Sci USA 107(50):21499–21504
Thurman RE et al (2012) The accessible chromatin landscape of the human genome. Nature 489(7414):75–82
Petersen B et al (2009) A generic method for assignment of reliability scores applied to solvent accessibility predictions. BMC Struct Biol 9:51
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26(1):139–140
Herz HM et al (2012) Enhancer-associated H3K4 monomethylation by Trithorax-related, the Drosophila homolog of mammalian Mll3/Mll4. Genes Dev 26(23):2604–2620
Subramanian K et al (2008) Regulation of estrogen receptor alpha by the SET7 lysine methyltransferase. Mol Cell 30(3):336–347
Couture JF et al (2006) Structural basis for the methylation site specificity of SET7/9. Nat Struct Mol Biol 13(2):140–146
Li Y et al (2008) Role of the histone H3 lysine 4 methyltransferase, SET7/9, in the regulation of NF-kappaB-dependent inflammatory genes. Relevance to diabetes and inflammation. J Biol Chem 283(39):26771–26781
Lamonica JM, Vakoc CR, Blobel GA (2006) Acetylation of GATA-1 is required for chromatin occupancy. Blood 108(12):3736–3738
Barski A et al (2007) High-resolution profiling of histone methylations in the human genome. Cell 129(4):823–837
Gaughan L et al (2011) Regulation of the androgen receptor by SET9-mediated methylation. Nucleic Acids Res 39(4):1266–1279
Lehnertz B et al (2011) p53-dependent transcription and tumor suppression are not affected in Set7/9-deficient mice. Mol Cell 43(4):673–680
Deering TG et al (2009) Methyltransferase Set7/9 maintains transcription and euchromatin structure at islet-enriched genes. Diabetes 58(1):185–193
Sun G et al (2010) Epigenetic histone methylation modulates fibrotic gene expression. J Am Soc Nephrol 21(12):2069–2080
Tao Y et al (2011) The histone methyltransferase Set7/9 promotes myoblast differentiation and myofibril assembly. J Cell Biol 194(4):551–565
Masatsugu T, Yamamoto K (2009) Multiple lysine methylation of PCAF by Set9 methyltransferase. Biochem Biophys Res Commun 381(1):22–26
Sikorski K et al (2012) STAT1 as a central mediator of IFNgamma and TLR4 signal integration in vascular dysfunction. JAKSTAT 1(4):241–249
Sikorski K et al (2011) STAT1 as a novel therapeutical target in pro-atherogenic signal integration of IFNgamma, TLR4 and IL-6 in vascular disease. Cytokine Growth Factor Rev 22(4):211–219
Sikorski K et al (2011) STAT1-mediated signal integration between IFNgamma and LPS leads to increased EC and SMC activation and monocyte adhesion. Am J Physiol Cell Physiol 300(6):C1337–C1344
Miao F et al (2014) Evaluating the role of epigenetic histone modifications in the metabolic memory of type 1 diabetes. Diabetes 63(5):1748–1762
Ko S et al (2011) Lysine methylation and functional modulation of androgen receptor by Set9 methyltransferase. Mol Endocrinol 25(3):433–444
Keating ST, El-Osta A (2012) Chromatin modifications associated with diabetes. J Cardiovasc Transl Res 5(4):399–412
Keating ST, El-Osta A (2013) Glycemic memories and the epigenetic component of diabetic nephropathy. Curr Diab Rep 13(4):574–581
Allis CD et al (2007) New nomenclature for chromatin-modifying enzymes. Cell 131(4):633–636
Glozak MA et al (2005) Acetylation and deacetylation of non-histone proteins. Gene 363:15–23
GENCODE v14 annotation URL ftp.sanger.ac.uk/pub/gencode/release_14/gencode.v14.annotation.gtf.gz
Acknowledgments
The authors are thankful for expert bioinformatic support by Antony Kaspi and Dr. Ross Lazarus. The authors acknowledge grant and fellowship support from the Juvenile Diabetes Research Foundation International, the Diabetes Australia Research Trust, the National Health and Medical Research Council (NHMRC) and the National Heart Foundation of Australia. STK is supported by an Australian Postgraduate Award. AE-O is a senior research fellow supported by the NHMRC. Supported in part by the Victorian Government’s Operational Infrastructure Support program.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
18_2014_1651_MOESM2_ESM.png
Supplemental Fig.S1 shRNA knockdown of Set7 confers reproducible and widespread changes in gene expression measured by mRNA-Seq. (A) Shows the extent to which the shRNA reduced the expression of SETD7 measured across three independent biological replicates in mRNA-Seq expressed as read counts per million (CPM) **p < 0.01. (B) Multi-dimensional scaling plot shows separation of control and knockdown samples. (C) Smear plot showing widespread changes in gene expression conferred by specific knockdown of Set7 expression. False discovery rate (FDR). (PNG 298 kb)
18_2014_1651_MOESM3_ESM.jpg
Supplemental Fig.S2 Genome-wide changes to mRNA expression associated with Set7 are regulated by H3K4m1 and transcription factor-dependent mechanisms. The outer ring depicts the positions of genes down-regulated by Set7KD along individual chromosomes. Red bars depicting transcriptional repression events identified by mRNA-Seq show widespread changes in Set7KD cells. The association of gene expression with changes to H3K4m1 patterns determined by ChIP-Seq is shown in green. The abundance of genes associated with transactivity of characterized Set7 substrate NFκB-p65 represented by blue bars partly overlaps the connectivity of putative novel Set7 substrate STAT1 (orange) to transcriptional deregulation. (JPEG 3738 kb)
Rights and permissions
About this article
Cite this article
Keating, S.T., Ziemann, M., Okabe, J. et al. Deep sequencing reveals novel Set7 networks. Cell. Mol. Life Sci. 71, 4471–4486 (2014). https://doi.org/10.1007/s00018-014-1651-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-014-1651-y