Abstract
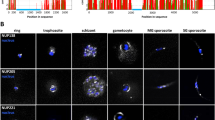
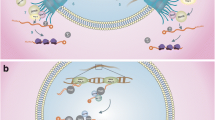
The nuclear pore is a key structure in eukaryotes regulating nuclear-cytoplasmic transport as well as a wide range of cellular processes. Here, we report the characterization of the first Toxoplasma gondii nuclear pore protein, named TgNup302, which appears to be the orthologue of the mammalian Nup98-96 protein. We produced a conditional knock-down mutant that expresses TgNup302 under the control of an inducible tetracycline-regulated promoter. Under ATc treatment, a substantial decrease of TgNup302 protein in inducible knock-down (iKD) parasites was observed, causing a delay in parasite proliferation. Moreover, the nuclear protein TgENO2 was trapped in the cytoplasm of ATc-treated mutants, suggesting that TgNup302 is involved in nuclear transport. Fluorescence in situ hybridization revealed that TgNup302 is essential for 18S RNA export from the nucleus to the cytoplasm, while global mRNA export remains unchanged. Using an affinity tag purification combined with mass spectrometry, we identified additional components of the nuclear pore complex, including proteins potentially interacting with chromatin. Furthermore, reverse immunoprecipitation confirmed their interaction with TgNup302, and structured illuminated microscopy confirmed the NPC localization of some of the TgNup302-interacting proteins. Intriguingly, facilitates chromatin transcription complex (FACT) components were identified, suggesting the existence of an NPC-chromatin interaction in T. gondii. Identification of TgNup302-interacting proteins also provides the first glimpse at the NPC structure in Apicomplexa, suggesting a structural conservation of the NPC components between distant eukaryotes.








Similar content being viewed by others
Abbreviations
- ATc:
-
AnhydroTetraCycline
- Co-IP:
-
Co-immunoprecipitation
- FACT:
-
Facilitates chromatin transcription
- FISH:
-
Fluorescence in situ hybridization
- IFA:
-
Immunofluorescence assay
- iKD:
-
Conditional knock-down
- NPC:
-
Nuclear pore complex
- NUP:
-
Nucleoporin
- RNA-Seq:
-
RNA-sequencing
- Sg RNA:
-
Single guide RNA
- SIM:
-
Structured illumination microscopy
- Tg:
-
Toxoplasma gondii
References
Kim K, Weiss LM (2004) Toxoplasma gondii: the model apicomplexan. Int J Parasitol 34:423–432
Behnke MS, Radke JB, Smith AT, Sullivan WJ, White MW (2008) The transcription of bradyzoite genes in Toxoplasma gondii is controlled by autonomous promoter elements. Mol Microbiol 68:1502–1518. doi:10.1111/j.1365-2958.2008.06249.x
Behnke MS, Wootton JC, Lehmann MM, Radke JB, Lucas O, Nawas J, Sibley LD, White MW (2010) Coordinated progression through two subtranscriptomes underlies the tachyzoite cycle of Toxoplasma gondii. PLoS One 5:e12354. doi:10.1371/journal.pone.0012354
Sullivan WJ, Hakimi MA (2006) Histone mediated gene activation in Toxoplasma gondii. Mol Biochem Parasitol 148:109–116
Gissot M, Kelly KA, Ajioka JW, Greally JM, Kim K (2007) Epigenomic modifications predict active promoters and gene structure in Toxoplasma gondii. PLoS Pathog 3:e77
Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, Sali A, Rout MP (2007) The molecular architecture of the nuclear pore complex. Nature 450:695–701. doi:10.1038/nature06405
Capelson M, Hetzer MW (2009) The role of nuclear pores in gene regulation, development and disease. EMBO Rep 10:697–705. doi:10.1038/embor.2009.147
Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT (2000) The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol 148:635–651
Vasu SK, Forbes DJ (2001) Nuclear pores and nuclear assembly. Curr Opin Cell Biol 13:363–375
Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ (2002) Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol 158:915–927. doi:10.1083/jcb.200206106
DeGrasse JA, DuBois KN, Devos D, Siegel TN, Sali A, Field MC, Rout MP, Chait BT (2009) Evidence for a shared nuclear pore complex architecture that is conserved from the last common eukaryotic ancestor. Mol Cell Proteom 8:2119–2130. doi:10.1074/mcp.M900038-MCP200
Obado SO, Brillantes M, Uryu K, Zhang W, Ketaren NE, Chait BT, Field MC, Rout MP (2016) Interactome mapping reveals the evolutionary history of the nuclear pore complex. PLoS Biol 14:e1002365. doi:10.1371/journal.pbio.1002365
R.Y.H. Lim, Fahrenkrog B, Köser J, Schwarz-Herion K, Deng J (2007) U. Aebi, Nanomechanical Basis of Selective Gating by the Nuclear Pore Complex. Science 318:640–643. doi:10.1126/science.1145980
Weiner A, Dahan-Pasternak N, Shimoni E, Shinder V, von Huth P, Elbaum M, Dzikowski R (2011) 3D nuclear architecture reveals coupled cell cycle dynamics of chromatin and nuclear pores in the malaria parasite Plasmodium falciparum. Cell Microbiol 13:967–977. doi:10.1111/j.1462-5822.2011.01592.x
Dahan-Pasternak N, Nasereddin A, Kolevzon N, Pe’er M, Wong W, Shinder V, Turnbull L, Whitchurch CB, Elbaum M, Gilberger TW, Yavin E, Baum J, Dzikowski R (2013) PfSec13 is an unusual chromatin-associated nucleoporin of Plasmodium falciparum that is essential for parasite proliferation in human erythrocytes. J Cell Sci 126:3055–3069. doi:10.1242/jcs.122119
Sheiner L, Demerly JL, Poulsen N, Beatty WL, Lucas O, Behnke MS, White MW, Striepen B (2011) A systematic screen to discover and analyze apicoplast proteins identifies a conserved and essential protein import factor. PLoS Pathog 7:e1002392. doi:10.1371/journal.ppat.1002392
Dzierszinski F, Mortuaire M, Dendouga N, Popescu O, Tomavo S (2001) Differential expression of two plant-like enolases with distinct enzymatic and antigenic properties during stage conversion of the protozoan parasite Toxoplasma gondii. J Mol Biol 309:1017–1027. doi:10.1006/jmbi.2001.4730
Gissot M, Walker R, Delhaye S, Huot L, Hot D, Tomavo S (2012) Toxoplasma gondii chromodomain protein 1 binds to heterochromatin and colocalises with centromeres and telomeres at the nuclear periphery. PLoS One 7:e32671. doi:10.1371/journal.pone.0032671
Olguin-Lamas A, Madec E, Hovasse A, Werkmeister E, Callebaut I, Slomianny C, Delhaye S, Mouveaux T, Schaeffer-Reiss C, Van Dorsselaer A, Tomavo S (2011) A novel Toxoplasma gondii nuclear factor TgNF3 is a dynamic chromatin-associated component, modulator of nucleolar architecture and parasite virulence. PLoS Pathog. 7:e1001328. doi:10.1371/journal.ppat.1001328
Gissot M, Walker R, Delhaye S, Alayi TD, Huot L, Hot D, Callebaut I, Schaeffer-Reiss C, Dorsselaer AV, Tomavo S (2013) Toxoplasma gondii Alba proteins are involved in translational control of gene expression. J Mol Biol 425:1287–1301. doi:10.1016/j.jmb.2013.01.039
Thompson J (2002) In situ detection of RNA in blood- and mosquito-stage malaria parasites. Methods Mol Med 72:225–233. doi:10.1385/1-59259-271-6:225
Shen B, Brown KM, Lee TD, Sibley LD (2014) Efficient gene disruption in diverse strains of Toxoplasma gondii using CRISPR/CAS9. MBio 5:e01114–01114. doi:10.1128/mBio.01114-14
Miguet L, Béchade G, Fornecker L, Zink E, Felden C, Gervais C, Herbrecht R, Van Dorsselaer A, van Dorsselaer A, Mauvieux L, Sanglier-Cianferani S (2009) Proteomic analysis of malignant B-cell derived microparticles reveals CD148 as a potentially useful antigenic biomarker for mantle cell lymphoma diagnosis. J Proteome Res 8:3346–3354. doi:10.1021/pr801102c
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi:10.1093/bioinformatics/btp324
Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14:R36. doi:10.1186/gb-2013-14-4-r36
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. doi:10.1038/nmeth.1923
Anders S, Pyl PT, Huber W (2015) HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics 31:166–169. doi:10.1093/bioinformatics/btu638
Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, J.Y.H. Yang, Zhang J (2004) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 5:R80. doi:10.1186/gb-2004-5-10-r80
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. doi:10.1093/bioinformatics/btp616
Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11:R106. doi:10.1186/gb-2010-11-10-r106
Tamura K, Fukao Y, Iwamoto M, Haraguchi T, Hara-Nishimura I (2010) Identification and characterization of nuclear pore complex components in Arabidopsis thaliana. Plant Cell 22:4084–4097. doi:10.1105/tpc.110.079947
Tamura K, Hara-Nishimura I (2013) The molecular architecture of the plant nuclear pore complex. J Exp Bot 64:823–832. doi:10.1093/jxb/ers258
Fontoura BM, Blobel G, Matunis MJ (1999) A conserved biogenesis pathway for nucleoporins: proteolytic processing of a 186-kilodalton precursor generates Nup98 and the novel nucleoporin, Nup96. J Cell Biol 144:1097–1112
Ferguson DJP, Parmley SF, Tomavo S (2002) Evidence for nuclear localisation of two stage-specific isoenzymes of enolase in Toxoplasma gondii correlates with active parasite replication. Int J Parasitol 32:1399–1410
Griffis ER, Xu S, Powers MA (2003) Nup98 localizes to both nuclear and cytoplasmic sides of the nuclear pore and binds to two distinct nucleoporin subcomplexes. Mol Biol Cell 14:600–610. doi:10.1091/mbc.E02-09-0582
Enninga J, Levay A, Fontoura BMA (2003) Sec13 shuttles between the nucleus and the cytoplasm and stably interacts with Nup96 at the nuclear pore complex. Mol Cell Biol 23:7271–7284
Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. doi:10.1038/nprot.2015.053
Pritchard CE, Fornerod M, Kasper LH, van Deursen JM (1999) RAE1 is a shuttling mRNA export factor that binds to a GLEBS-like NUP98 motif at the nuclear pore complex through multiple domains. J Cell Biol 145:237–254
Guizetti J, Martins RM, Guadagnini S, Claes A, Scherf A (2013) Nuclear pores and perinuclear expression sites of var and ribosomal DNA genes correspond to physically distinct regions in Plasmodium falciparum. Eukaryot Cell 12:697–702. doi:10.1128/EC.00023-13
Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, Jaenisch R (2013) One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 153:910–918. doi:10.1016/j.cell.2013.04.025
Fabre E, Boelens WC, Wimmer C, Mattaj IW, Hurt EC (1994) Nup145p is required for nuclear export of mRNA and binds homopolymeric RNA in vitro via a novel conserved motif. Cell 78:275–289
Dockendorff TC, Heath CV, Goldstein AL, Snay CA, Cole CN (1997) C-terminal truncations of the yeast nucleoporin Nup145p produce a rapid temperature-conditional mRNA export defect and alterations to nuclear structure. Mol Cell Biol 17:906–920
Powers MA, Forbes DJ, Dahlberg JE, Lund E (1997) The vertebrate GLFG nucleoporin, Nup98, is an essential component of multiple RNA export pathways. J Cell Biol 136:241–250
Radu A, Blobel G, Moore MS (1995) Identification of a protein complex that is required for nuclear protein import and mediates docking of import substrate to distinct nucleoporins. Proc Natl Acad Sci USA 92:1769–1773
Radu A, Moore MS, Blobel G (1995) The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell 81:215–222
Frankel MB, Knoll LJ (2009) The ins and outs of nuclear trafficking: unusual aspects in apicomplexan parasites. DNA Cell Biol 28:277–284. doi:10.1089/dna.2009.0853
Brooks CF, Francia ME, Gissot M, Croken MM, Kim K, Striepen B (2011) Toxoplasma gondii sequesters centromeres to a specific nuclear region throughout the cell cycle. Proc Natl Acad Sci USA 108:3767–3772. doi:10.1073/pnas.1006741108
Lemaître C, Bickmore WA (2015) Chromatin at the nuclear periphery and the regulation of genome functions. Histochem Cell Biol 144:111–122. doi:10.1007/s00418-015-1346-y
Liang Y, Hetzer MW (2011) Functional interactions between nucleoporins and chromatin. Curr Opin Cell Biol 23:65–70. doi:10.1016/j.ceb.2010.09.008
Brown CR, Kennedy CJ, Delmar VA, Forbes DJ, Silver PA (2008) Global histone acetylation induces functional genomic reorganization at mammalian nuclear pore complexes. Genes Dev 22:627–639. doi:10.1101/gad.1632708
Bougdour A, Maubon D, Baldacci P, Ortet P, Bastien O, Bouillon A, Barale J-C, Pelloux H, Ménard R, Hakimi M-A (2009) Drug inhibition of HDAC3 and epigenetic control of differentiation in Apicomplexa parasites. J Exp Med 206:953–966. doi:10.1084/jem.20082826
Reinberg D, Sims RJ (2006) de FACTo nucleosome dynamics. J Biol Chem 281:23297–23301. doi:10.1074/jbc.R600007200
Tous C, Rondón AG, García-Rubio M, González-Aguilera C, Luna R, Aguilera A (2011) A novel assay identifies transcript elongation roles for the Nup84 complex and RNA processing factors. EMBO J 30:1953–1964. doi:10.1038/emboj.2011.109
Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, J.L.Y. Koh, Toufighi K, Mostafavi S, Prinz J, St Onge RP, VanderSluis B, Makhnevych T, Vizeacoumar FJ, Alizadeh S, Bahr S, Brost RL, Chen Y, Cokol M, Deshpande R, Li Z, Lin Z-Y, Liang W, Marback M, Paw J, San Luis B-J, Shuteriqi E, A.H.Y. Tong, van Dyk N, Wallace IM, Whitney JA, Weirauch MT, Zhong G, Zhu H, Houry WA, Brudno M, Ragibizadeh S, Papp B, Pál C, Roth FP, Giaever G, Nislow C, Troyanskaya OG, Bussey H, Bader GD, Gingras A-C, Morris QD, Kim PM, Kaiser CA, Myers CL, Andrews BJ, Boone C (2010) The genetic landscape of a cell. Science 327:425–431. doi:10.1126/science.1180823
Burckin T, Nagel R, Mandel-Gutfreund Y, Shiue L, Clark TA, Chong J-L, Chang T-H, Squazzo S, Hartzog G, Ares M (2005) Exploring functional relationships between components of the gene expression machinery. Nat Struct Mol Biol 12:175–182. doi:10.1038/nsmb891
Hautbergue GM, Hung M-L, Walsh MJ, A.P.L. Snijders, Chang C-T, Jones R, Ponting CP, Dickman MJ, Wilson SA (2009) UIF, a new mRNA export adaptor that works together with REF/ALY, requires FACT for recruitment to mRNA. Curr Biol 19:1918–1924. doi:10.1016/j.cub.2009.09.041
Acknowledgements
The authors would like to thank Dr. Valerie Doye for helpful discussions and Dr. R. Walker for critically reading the manuscript. We also thank Ludovic Huot for checking the integrity of RNA samples, Etienne Dewailly for electronic microscopy, Antonino Bongiovanni for his help with microscopy data analyses, and Quentin Deveuve for phylogenetic tree recommendations. The authors also thank the BioImaging Center Lille for access to instruments. This work was supported by Centre National de la Recherche Scientifique (CNRS), Institut National de la Santé et de la Recherche Médicale (INSERM), grants from the French National Research Agency (ANR) [Grant Number ANR-13-JSV3-0006-01 to MG and ANR-11-LABX-0024 to Plateforme Protéomique et Peptides Modifiés (P3M)], the Fonds Européen de Développement Economique Régionale (13003300-42405 Labex Parafrap to P3M) and the Métropole Européenne de Lille (MEL).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
18_2017_2459_MOESM3_ESM.xls
Table S3 Mass-spectrometry results. The proteins highlighted in orange met the following criteria: less than 1 peptide in the control experiment and present in the two IP experiments (XLS 226 KB)
18_2017_2459_MOESM4_ESM.pdf
Fig. S1 T. gondii TgNup302 is evolutionarily conserved among Eukaryote. Phylogenetic tree of T. gondii TgNup302 homologues based on ClustalW alignment of sequences identified by BLASTp searches using the entire sequence of TgTgNup302 gene against 15 apicomplexan parasites (brown), 5 Fungi (green), 3 Eumetazoa (blue) and 3 Plantae (purple). The tree was reconstructed by maximum likelihood (ML) analyses with MEGA6 software. Five hundred bootstrap pseudo-replicates were used to give statistical support to the clades of the maximum likelihood topology. Scale bar reflects number of substitutions per site. Numbers in the nodes of the tree reflect the percentage of bootstrap replicates supporting each node (PDF 39 KB)
18_2017_2459_MOESM5_ESM.pdf
Fig. S2 Construction of the TgNup302 iKD strain. (A) Schematic of the genetic approach used to produce the conditional knock-down strain by a promoter replacement strategy. After promoter replacement, the expression of TgNup302 is under the control of anhydrotetracyline (ATc). The TgNup302 gene was HA-tagged at its 5’ (Top panel). (B) PCR was used to confirm the correct integration of the plasmid and the creation of the recombinant locus using primer pairs i and ii (sequences are in TableS1 of supplementary data). (C) Double HA-tagged (N-terminal) / Myc-tagged (C-terminal) strain for the TgNup302 gene was produced by introducing a myc tag at the 3’ end of the gene (PDF 54 KB)
18_2017_2459_MOESM6_ESM.pdf
Fig. S3 PolyA+ RNA were mostly cytoplasmic in the parental and iKD strains. RNA FISH was performed on intracellular parasites. Parasites of the parental and TgNup302 iKD strains were hybridized with Cy3-labeled polyA+ oligonucleotides primers (red), and the nuclear DNA was labeled with DAPI (blue) (PDF 284 KB)
18_2017_2459_MOESM7_ESM.pdf
Fig. S4 Conditional expression of TgNup302 has no impact on TgChromo1 and TgNF3 localization. Endogenous TgChromo1 and TgNF3 were labeled with the mouse monoclonal anti-Myc and 488-nm Alexa goat secondary antibody in parental and TgNup302 iKD strains with or without ATc treatment for 48hr (PDF 867 KB)
18_2017_2459_MOESM8_ESM.pdf
Fig. S5 Ultrastructure of Toxoplasma gondii. Intracellular T. gondii tachyzoite showing the nucleus (N) for the parental RHΔKu80 TaTi and iKD TgNup302 strains with or without ATc treatment. Bar=500 nm (PDF 1529 KB)
18_2017_2459_MOESM9_ESM.pdf
Fig. S6 Chromosomal distribution of genes differentially regulated in the TgNUP1 iKD strain. The chromosomal position and distribution of genes that were identified as differentially expressed, upregulated (top panel), and downregulated (bottom panel), in TgNup302 iKD after an ATc treatment for 48hr (PDF 777 KB)
18_2017_2459_MOESM10_ESM.pdf
Fig. S7 Peptides identified for TgNup302. Peptides recovered from the immunoprecipitation of TgNup302 and mapped onto the TgNup302 sequence are highlighted in yellow. The autocatalytic domain sequence is underlined (PDF 132 KB)
18_2017_2459_MOESM11_ESM.pdf
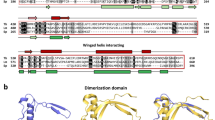
Fig. S8 Predicted secondary structure features, fold, and location for validated TgNups. The horizontal black line represents the polypeptide length of the proteins. The y-axis indicates the confidence score of the predicted secondary structure element. Predicted α-helices are indicated in blue, predicted β-sheets in orange, and predicted coiled-coil regions are in red arrows. The green arrows indicate FG repeats (PDF 353 KB)
18_2017_2459_MOESM12_ESM.pdf
Fig. S9 Identified proteins interact with TgNup302. (a) 5 µl of nuclear extract (from ~500x106 parasites) conserved before the immunoprecipitation (inputs) for the TgFACT140 (line 2), TgNup134 (line 3), TgNup129 (line 4) and TgNup407 (line 5) C-terminally myc-tagged proteins in the TgNup302-HA iKD strain were analyzed by Western blot. The immunoblot was probed with anti-HA antibody to detect the presence of the TgNup302-HA protein in each strain before immunoprecipitation. (b) Immunoblot was reprobed with an anti-myc antibody to detect the myc-tagged proteins: TgFACT140 (134 kDa), TgNup129 (129 kDa), TgNup134 (134 kDa), and TgNup407 (115 kDa) (PDF 91 KB)
18_2017_2459_MOESM13_ESM.pdf
Fig. S10 New components of the T. gondii nuclear pore. Each potential partner was tagged using a Myc-tag in the TgNup302-HA iKD strain. Endogenous TgNup302 iKD was labeled with the rabbit monoclonal anti-HA (in red) and endogenous TgFACT140, TgNup129 were labeled with the mouse monoclonal anti-Myc (in green) antibody with or without ATc after 48hr of growth (PDF 446 KB)
18_2017_2459_MOESM14_ESM.pdf
Fig. S11 Transfection efficiency for the Crisp-Cas9 screening. For each construction, the percentage of vacuole with a positive GFP expression was monitored to determine the transfection efficiency at 24 h after electroporation, revealing that ~30 to 70% of cells received the plasmid. TgAlba1 is a negative control (PDF 49 KB)
Rights and permissions
About this article
Cite this article
Courjol, F., Mouveaux, T., Lesage, K. et al. Characterization of a nuclear pore protein sheds light on the roles and composition of the Toxoplasma gondii nuclear pore complex. Cell. Mol. Life Sci. 74, 2107–2125 (2017). https://doi.org/10.1007/s00018-017-2459-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-017-2459-3