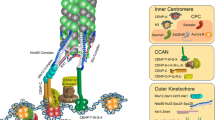
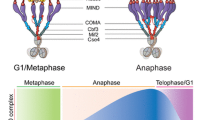
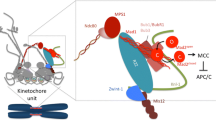
Abstract
Faithful chromosome segregation during mitosis in eukaryotes requires attachment of the kinetochore, a large protein complex assembled on the centromere of each chromosome, to the spindle microtubules. The kinetochore is a structural interface for the microtubule attachment and provides molecular surveillance mechanisms that monitor and ensure the precise microtubule attachment as well, including error correction and spindle assembly checkpoint. During mitotic progression, the kinetochore undergoes dynamic morphological changes that are observable through electron microscopy as well as through fluorescence microscopy. These structural changes might be associated with the kinetochore function. In this review, we summarize how the dynamics of kinetochore morphology are associated with its functions and discuss recent findings on the switching of protein interaction networks in the kinetochore during cell cycle progression.







Similar content being viewed by others
References
Cheeseman IM (2014) The kinetochore. Cold Spring Harb Perspect Biol 6:a015826–a015826. https://doi.org/10.1101/cshperspect.a015826
Musacchio A, Desai A (2017) A molecular view of kinetochore assembly and function. Biology 5(6):5. https://doi.org/10.3390/biology6010005
Fukagawa T, Earnshaw WC (2014) The centromere: chromatin foundation for the kinetochore machinery. Dev Cell 30:496–508. https://doi.org/10.1016/j.devcel.2014.08.016
Musacchio A (2015) The molecular biology of spindle assembly checkpoint signaling dynamics. Curr Biol 25:R1002–R1018. https://doi.org/10.1016/j.cub.2015.08.051
Sacristan C, Kops GJPL (2015) Joined at the hip: kinetochores, microtubules, and spindle assembly checkpoint signaling. Trends Cell Biol 25:21–28. https://doi.org/10.1016/j.tcb.2014.08.006
Sarangapani KK, Asbury CL (2014) Catch and release: how do kinetochores hook the right microtubules during mitosis? Trends Genet 30:150–159. https://doi.org/10.1016/j.tig.2014.02.004
Nagpal H, Fukagawa T (2016) Kinetochore assembly and function through the cell cycle. Chromosoma 125:645–659. https://doi.org/10.1007/s00412-016-0608-3
Pesenti ME, Weir JR, Musacchio A (2016) Progress in the structural and functional characterization of kinetochores. Curr Opin Struct Biol 37:152–163. https://doi.org/10.1016/j.sbi.2016.03.003
Thrower DA, Jordan MA, Wilson L (1996) Modulation of CENP-E organization at kinetochores by spindle microtubule attachment. Cell Motil Cytoskeleton 35:121–133. https://doi.org/10.1002/(SICI)1097-0169(1996)35:2%3c121:AID-CM5%3e3.0.CO;2-D
Rieder CL (1982) The formation, structure, and composition of the mammalian kinetochore and kinetochore fiber. Int Rev Cytol 79:1–58. https://doi.org/10.1016/s0074-7696(08)61672-1
McEwen BF, Hsieh CE, Mattheyses AL, Rieder CL (1998) A new look at kinetochore structure in vertebrate somatic cells using high-pressure freezing and freeze substitution. Chromosoma 107:366–375. https://doi.org/10.1007/s004120050320
Hoffman DB, Pearson CG, Yen TJ et al (2001) Microtubule-dependent changes in assembly of microtubule motor proteins and mitotic spindle checkpoint proteins at PtK1 kinetochores. Mol Biol Cell 12:1995–2009. https://doi.org/10.1091/mbc.12.7.1995
Wynne DJ, Funabiki H (2015) Kinetochore function is controlled by a phospho-dependent coexpansion of inner and outer components. J Cell Biol 210:899–916. https://doi.org/10.1083/jcb.201506020
Krenn V, Musacchio A (2015) The Aurora B kinase in chromosome bi-orientation and spindle checkpoint signaling. Front Oncol 5:225. https://doi.org/10.3389/fonc.2015.00225
Lampson MA, Cheeseman IM (2011) Sensing centromere tension: Aurora B and the regulation of kinetochore function. Trends Cell Biol 21:133–140. https://doi.org/10.1016/j.tcb.2010.10.007
Maresca TJ, Salmon ED (2010) Welcome to a new kind of tension: translating kinetochore mechanics into a wait-anaphase signal. J Cell Sci 123:825–835. https://doi.org/10.1242/jcs.064790
Hara M, Fukagawa T (2017) Critical foundation of the kinetochore: the constitutive centromere-associated network (CCAN). Prog Mol Subcell Biol 56:29–57. https://doi.org/10.1007/978-3-319-58592-5_2
Varma D, Salmon ED (2012) The KMN protein network—chief conductors of the kinetochore orchestra. J Cell Sci 125:5927–5936. https://doi.org/10.1242/jcs.093724
Monda JK, Cheeseman IM (2018) The kinetochore-microtubule interface at a glance. J Cell Sci 131:jcs214577. https://doi.org/10.1242/jcs.214577
Hara M, Ariyoshi M, Okumura E-I et al (2018) Multiple phosphorylations control recruitment of the KMN network onto kinetochores. Nat Cell Biol 20:1378–1388. https://doi.org/10.1038/s41556-018-0230-0
Watanabe R, Hara M, Okumura E-I et al (2019) CDK1-mediated CENP-C phosphorylation modulates CENP-A binding and mitotic kinetochore localization. J Cell Biol 218:4042–4062. https://doi.org/10.1083/jcb.201907006
McKinley KL, Cheeseman IM (2016) The molecular basis for centromere identity and function. Nat Rev Mol Cell Biol 17:16–29. https://doi.org/10.1038/nrm.2015.5
Brinkley BR, Stubblefield E (1966) The fine structure of the kinetochore of a mammalian cell in vitro. Chromosoma 19:28–43. https://doi.org/10.1007/bf00332792
Jokelainen PT (1967) The ultrastructure and spatial organization of the metaphase kinetochore in mitotic rat cells. J Ultrastruct Res 19:19–44. https://doi.org/10.1016/s0022-5320(67)80058-3
Rieder CL (1981) The structure of the cold-stable kinetochore fiber in metaphase PtK1 cells. Chromosoma 84:145–158. https://doi.org/10.1007/bf00293368
McEwen BF, Arena JT, Frank J, Rieder CL (1993) Structure of the colcemid-treated PtK1 kinetochore outer plate as determined by high voltage electron microscopic tomography. J Cell Biol 120:301–312. https://doi.org/10.1083/jcb.120.2.301
McEwen BF, Dong Y (2010) Contrasting models for kinetochore microtubule attachment in mammalian cells. Cell Mol Life Sci 67:2163–2172. https://doi.org/10.1007/s00018-010-0322-x
Maiato H, DeLuca J, Salmon ED, Earnshaw WC (2004) The dynamic kinetochore–microtubule interface. J Cell Sci 117:5461–5477. https://doi.org/10.1242/jcs.01536
Cheeseman IM, Chappie JS, Wilson-Kubalek EM, Desai A (2006) The conserved KMN network constitutes the core microtubule-binding site of the kinetochore. Cell 127:983–997. https://doi.org/10.1016/j.cell.2006.09.039
Cheeseman IM, Desai A (2008) Molecular architecture of the kinetochore–microtubule interface. Nat Rev Mol Cell Biol 9:33–46. https://doi.org/10.1038/nrm2310
Foley EA, Kapoor TM (2013) Microtubule attachment and spindle assembly checkpoint signalling at the kinetochore. Nat Rev Mol Cell Biol 14:25–37. https://doi.org/10.1038/nrm3494
DeLuca JG, Dong Y, Hergert P et al (2005) Hec1 and nuf2 are core components of the kinetochore outer plate essential for organizing microtubule attachment sites. Mol Biol Cell 16:519–531. https://doi.org/10.1091/mbc.e04-09-0852
Ciferri C, Pasqualato S, Screpanti E et al (2008) Implications for kinetochore–microtubule attachment from the structure of an engineered Ndc80 complex. Cell 133:427–439. https://doi.org/10.1016/j.cell.2008.03.020
DeLuca JG, Gall WE, Ciferri C et al (2006) Kinetochore microtubule dynamics and attachment stability are regulated by Hec1. Cell 127:969–982. https://doi.org/10.1016/j.cell.2006.09.047
Wei RR, Al-Bassam J, Harrison SC (2007) The Ndc80/HEC1 complex is a contact point for kinetochore–microtubule attachment. Nat Struct Mol Biol 14:54–59. https://doi.org/10.1038/nsmb1186
Cassimeris L, Rieder CL, Rupp G, Salmon ED (1990) Stability of microtubule attachment to metaphase kinetochores in PtK1 cells. J Cell Sci 96(Pt 1):9–15
Dong Y, Vanden Beldt KJ, Meng X et al (2007) The outer plate in vertebrate kinetochores is a flexible network with multiple microtubule interactions. Nat Cell Biol 9:516–522. https://doi.org/10.1038/ncb1576
Rieder CL, Alexander SP (1990) Kinetochores are transported poleward along a single astral microtubule during chromosome attachment to the spindle in newt lung cells. J Cell Biol 110:81–95. https://doi.org/10.1083/jcb.110.1.81
Yao X, Anderson KL, Cleveland DW (1997) The microtubule-dependent motor centromere-associated protein E (CENP-E) is an integral component of kinetochore corona fibers that link centromeres to spindle microtubules. J Cell Biol 139:435–447. https://doi.org/10.1083/jcb.139.2.435
Cooke CA, Schaar B, Yen TJ, Earnshaw WC (1997) Localization of CENP-E in the fibrous corona and outer plate of mammalian kinetochores from prometaphase through anaphase. Chromosoma 106:446–455. https://doi.org/10.1007/s004120050266
Magidson V, He J, Ault JG et al (2016) Unattached kinetochores rather than intrakinetochore tension arrest mitosis in taxol-treated cells. J Cell Biol 212:307–319. https://doi.org/10.1083/jcb.201412139
Johnson VL, Scott MIF, Holt SV et al (2004) Bub1 is required for kinetochore localization of BubR1, Cenp-E, Cenp-F and Mad2, and chromosome congression. J Cell Sci 117:1577–1589. https://doi.org/10.1242/jcs.01006
Rodriguez-Rodriguez J-A, Lewis C, McKinley KL et al (2018) Distinct roles of RZZ and Bub1-KNL1 in mitotic checkpoint signaling and kinetochore expansion. Curr Biol 28:3422–3429.e5. https://doi.org/10.1016/j.cub.2018.10.006
Pereira C, Reis RM, Gama JB et al (2018) Self-assembly of the RZZ complex into filaments drives kinetochore expansion in the absence of microtubule attachment. Curr Biol 28:3408–3421.e8. https://doi.org/10.1016/j.cub.2018.08.056
Magidson V, Paul R, Yang N et al (2015) Adaptive changes in the kinetochore architecture facilitate proper spindle assembly. Nat Cell Biol 17:1134–1144. https://doi.org/10.1038/ncb3223
Wynne DJ, Funabiki H (2016) Heterogeneous architecture of vertebrate kinetochores revealed by three-dimensional superresolution fluorescence microscopy. Mol Biol Cell 27:3395–3404. https://doi.org/10.1091/mbc.E16-02-0130
Sacristan C, Ahmad MUD, Keller J et al (2018) Dynamic kinetochore size regulation promotes microtubule capture and chromosome biorientation in mitosis. Nat Cell Biol 20:800–810. https://doi.org/10.1038/s41556-018-0130-3
Howell BJ, McEwen BF, Canman JC et al (2001) Cytoplasmic dynein/dynactin drives kinetochore protein transport to the spindle poles and has a role in mitotic spindle checkpoint inactivation. J Cell Biol 155:1159–1172. https://doi.org/10.1083/jcb.200105093
Wojcik E, Basto R, Serr M et al (2001) Kinetochore dynein: its dynamics and role in the transport of the Rough deal checkpoint protein. Nat Cell Biol 3:1001–1007. https://doi.org/10.1038/ncb1101-1001
Mische S, He Y, Ma L et al (2008) Dynein light intermediate chain: an essential subunit that contributes to spindle checkpoint inactivation. Mol Biol Cell 19:4918–4929. https://doi.org/10.1091/mbc.e08-05-0483
Sivaram MVS, Wadzinski TL, Redick SD et al (2009) Dynein light intermediate chain 1 is required for progress through the spindle assembly checkpoint. EMBO J 28:902–914. https://doi.org/10.1038/emboj.2009.38
Karess R (2005) Rod–Zw10–Zwilch: a key player in the spindle checkpoint. Trends Cell Biol 15:386–392. https://doi.org/10.1016/j.tcb.2005.05.003
Karess RE, Glover DM (1989) rough deal: a gene required for proper mitotic segregation in Drosophila. J Cell Biol 109:2951–2961. https://doi.org/10.1083/jcb.109.6.2951
Williams BC, Karr TL, Montgomery JM, Goldberg ML (1992) The Drosophila l(1)zw10 gene product, required for accurate mitotic chromosome segregation, is redistributed at anaphase onset. J Cell Biol 118:759–773. https://doi.org/10.1083/jcb.118.4.759
Starr DA, Williams BC, Li Z et al (1997) Conservation of the centromere/kinetochore protein ZW10. J Cell Biol 138:1289–1301. https://doi.org/10.1083/jcb.138.6.1289
Scaërou F, Starr DA, Piano F et al (2001) The ZW10 and Rough Deal checkpoint proteins function together in a large, evolutionarily conserved complex targeted to the kinetochore. J Cell Sci 114:3103–3114
Basto R, Gomes R, Karess RE (2000) Rough deal and Zw10 are required for the metaphase checkpoint in Drosophila. Nat Cell Biol 2:939–943. https://doi.org/10.1038/35046592
Chan GK, Jablonski SA, Starr DA et al (2000) Human Zw10 and ROD are mitotic checkpoint proteins that bind to kinetochores. Nat Cell Biol 2:944–947. https://doi.org/10.1038/35046598
Williams BC, Li Z, Liu S et al (2003) Zwilch, a new component of the ZW10/ROD complex required for kinetochore functions. Mol Biol Cell 14:1379–1391. https://doi.org/10.1091/mbc.e02-09-0624
Mosalaganti S, Keller J, Altenfeld A et al (2017) Structure of the RZZ complex and molecular basis of its interaction with Spindly. J Cell Biol 216:961–981. https://doi.org/10.1083/jcb.201611060
Griffis ER, Stuurman N, Vale RD (2007) Spindly, a novel protein essential for silencing the spindle assembly checkpoint, recruits dynein to the kinetochore. J Cell Biol 177:1005–1015. https://doi.org/10.1083/jcb.200702062
Chan YW, Fava LL, Uldschmid A et al (2009) Mitotic control of kinetochore-associated dynein and spindle orientation by human Spindly. J Cell Biol 185:859–874. https://doi.org/10.1083/jcb.200812167
Barisic M, Sohm B, Mikolcevic P et al (2010) Spindly/CCDC99 is required for efficient chromosome congression and mitotic checkpoint regulation. Mol Biol Cell 21:1968–1981. https://doi.org/10.1091/mbc.e09-04-0356
Gassmann R, Holland AJ, Varma D et al (2010) Removal of Spindly from microtubule-attached kinetochores controls spindle checkpoint silencing in human cells. Genes Dev 24:957–971. https://doi.org/10.1101/gad.1886810
Holland AJ, Reis RM, Niessen S et al (2015) Preventing farnesylation of the dynein adaptor Spindly contributes to the mitotic defects caused by farnesyltransferase inhibitors. Mol Biol Cell 26:1845–1856. https://doi.org/10.1091/mbc.E14-11-1560
Moudgil DK, Westcott N, Famulski JK et al (2015) A novel role of farnesylation in targeting a mitotic checkpoint protein, human Spindly, to kinetochores. J Cell Biol 208:881–896. https://doi.org/10.1083/jcb.201412085
Gassmann R, Essex A, Hu J-S et al (2008) A new mechanism controlling kinetochore–microtubule interactions revealed by comparison of two dynein-targeting components: SPDL-1 and the Rod/Zwilch/Zw10 complex. Genes Dev 22:2385–2399. https://doi.org/10.1101/gad.1687508
Zhang G, Lischetti T, Hayward DG, Nilsson J (2015) Distinct domains in Bub1 localize RZZ and BubR1 to kinetochores to regulate the checkpoint. Nat Commun 6:7162–7214. https://doi.org/10.1038/ncomms8162
Qian J, García-Gimeno MA, Beullens M et al (2017) An attachment-independent biochemical timer of the spindle assembly checkpoint. Mol Cell 68:715–730.e5. https://doi.org/10.1016/j.molcel.2017.10.011
Caldas GV, Lynch TR, Anderson R et al (2015) The RZZ complex requires the N-terminus of KNL1 to mediate optimal Mad1 kinetochore localization in human cells. Open Biol 5:150160. https://doi.org/10.1098/rsob.150160
Gama JB, Pereira C, Simões PA et al (2017) Molecular mechanism of dynein recruitment to kinetochores by the Rod–Zw10–Zwilch complex and Spindly. J Cell Biol 216:943–960. https://doi.org/10.1083/jcb.201610108
Silió V, McAinsh AD, Millar JB (2015) KNL1-Bubs and RZZ provide two separable pathways for checkpoint activation at human kinetochores. Dev Cell 35:600–613. https://doi.org/10.1016/j.devcel.2015.11.012
Kops GJPL, Kim Y, Weaver BAA et al (2005) ZW10 links mitotic checkpoint signaling to the structural kinetochore. J Cell Biol 169:49–60. https://doi.org/10.1083/jcb.200411118
Buffin E, Lefebvre C, Huang J et al (2005) Recruitment of Mad2 to the kinetochore requires the Rod/Zw10 complex. Curr Biol 15:856–861. https://doi.org/10.1016/j.cub.2005.03.052
Basto R, Scaerou F, Mische S et al (2004) In vivo dynamics of the rough deal checkpoint protein during Drosophila mitosis. Curr Biol 14:56–61. https://doi.org/10.1016/j.cub.2003.12.025
Waters JC, Mitchison TJ, Rieder CL, Salmon ED (1996) The kinetochore microtubule minus-end disassembly associated with poleward flux produces a force that can do work. Mol Biol Cell 7:1547–1558. https://doi.org/10.1091/mbc.7.10.1547
Maresca TJ, Salmon ED (2009) Intrakinetochore stretch is associated with changes in kinetochore phosphorylation and spindle assembly checkpoint activity. J Cell Biol 184:373–381. https://doi.org/10.1083/jcb.200808130
Uchida KSK, Takagaki K, Kumada K et al (2009) Kinetochore stretching inactivates the spindle assembly checkpoint. J Cell Biol 184:383–390. https://doi.org/10.1083/jcb.200811028
Wordeman L, Steuer ER, Sheetz MP, Mitchison T (1991) Chemical subdomains within the kinetochore domain of isolated CHO mitotic chromosomes. J Cell Biol 114:285–294. https://doi.org/10.1083/jcb.114.2.285
Schittenhelm RB, Heeger S, Althoff F et al (2007) Spatial organization of a ubiquitous eukaryotic kinetochore protein network in Drosophila chromosomes. Chromosoma 116:385–402. https://doi.org/10.1007/s00412-007-0103-y
Biggins S, Severin FF, Bhalla N et al (1999) The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev 13:532–544. https://doi.org/10.1101/gad.13.5.532
Tanaka TU, Rachidi N, Janke C et al (2002) Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell 108:317–329. https://doi.org/10.1016/s0092-8674(02)00633-5
Lampson MA, Renduchitala K, Khodjakov A, Kapoor TM (2004) Correcting improper chromosome-spindle attachments during cell division. Nat Cell Biol 6:232–237. https://doi.org/10.1038/ncb1102
Cimini D, Wan X, Hirel CB, Salmon ED (2006) Aurora kinase promotes turnover of kinetochore microtubules to reduce chromosome segregation errors. Curr Biol 16:1711–1718. https://doi.org/10.1016/j.cub.2006.07.022
Kelly AE, Funabiki H (2009) Correcting aberrant kinetochore microtubule attachments: an Aurora B-centric view. Curr Opin Cell Biol 21:51–58. https://doi.org/10.1016/j.ceb.2009.01.004
Adams RR, Maiato H, Earnshaw WC, Carmena M (2001) Essential roles of Drosophila inner centromere protein (INCENP) and aurora B in histone H3 phosphorylation, metaphase chromosome alignment, kinetochore disjunction, and chromosome segregation. J Cell Biol 153:865–880. https://doi.org/10.1083/jcb.153.4.865
Carvalho A, Carmena M, Sambade C et al (2003) Survivin is required for stable checkpoint activation in taxol-treated HeLa cells. J Cell Sci 116:2987–2998. https://doi.org/10.1242/jcs.00612
Honda R, Körner R, Nigg EA (2003) Exploring the functional interactions between Aurora B, INCENP, and survivin in mitosis. Mol Biol Cell 14:3325–3341. https://doi.org/10.1091/mbc.e02-11-0769
Gassmann R, Carvalho A, Henzing AJ et al (2004) Borealin: a novel chromosomal passenger required for stability of the bipolar mitotic spindle. J Cell Biol 166:179–191. https://doi.org/10.1083/jcb.200404001
Klein UR, Nigg EA, Gruneberg U (2006) Centromere targeting of the chromosomal passenger complex requires a ternary subcomplex of Borealin, Survivin, and the N-terminal domain of INCENP. Mol Biol Cell 17:2547–2558. https://doi.org/10.1091/mbc.e05-12-1133
Romano A, Guse A, Krascenicova I et al (2003) CSC-1: a subunit of the Aurora B kinase complex that binds to the survivin-like protein BIR-1 and the incenp-like protein ICP-1. J Cell Biol 161:229–236. https://doi.org/10.1083/jcb.200207117
Cooke CA, Heck MM, Earnshaw WC (1987) The inner centromere protein (INCENP) antigens: movement from inner centromere to midbody during mitosis. J Cell Biol 105:2053–2067. https://doi.org/10.1083/jcb.105.5.2053
Kaitna S, Mendoza M, Jantsch-Plunger V, Glotzer M (2000) Incenp and an aurora-like kinase form a complex essential for chromosome segregation and efficient completion of cytokinesis. Curr Biol 10:1172–1181. https://doi.org/10.1016/s0960-9822(00)00721-1
Sampath SC, Ohi R, Leismann O et al (2004) The chromosomal passenger complex is required for chromatin-induced microtubule stabilization and spindle assembly. Cell 118:187–202. https://doi.org/10.1016/j.cell.2004.06.026
Ambrosini G, Adida C, Altieri DC (1997) A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med 3:917–921. https://doi.org/10.1038/nm0897-917
Samejima K, Platani M, Wolny M et al (2015) The inner centromere protein (INCENP) coil is a single α-helix (SAH) domain that binds directly to microtubules and is important for chromosome passenger complex (CPC) localization and function in mitosis. J Biol Chem 290:21460–21472. https://doi.org/10.1074/jbc.M115.645317
Zaytsev AV, Segura-Peña D, Godzi M et al (2016) Bistability of a coupled Aurora B kinase-phosphatase system in cell division. Elife 5:e10644. https://doi.org/10.7554/eLife.10644
Wang E, Ballister ER, Lampson MA (2011) Aurora B dynamics at centromeres create a diffusion-based phosphorylation gradient. J Cell Biol 194:539–549. https://doi.org/10.1083/jcb.201103044
Liu D, Vader G, Vromans MJM et al (2009) Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science 323:1350–1353. https://doi.org/10.1126/science.1167000
Welburn JPI, Vleugel M, Liu D et al (2010) Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore–microtubule interface. Mol Cell 38:383–392. https://doi.org/10.1016/j.molcel.2010.02.034
DeLuca KF, Lens SMA, DeLuca JG (2011) Temporal changes in Hec1 phosphorylation control kinetochore–microtubule attachment stability during mitosis. J Cell Sci 124:622–634. https://doi.org/10.1242/jcs.072629
Cheeseman IM, Anderson S, Jwa M et al (2002) Phospho-regulation of kinetochore–microtubule attachments by the Aurora kinase Ipl1p. Cell 111:163–172. https://doi.org/10.1016/s0092-8674(02)00973-x
Guimaraes GJ, Dong Y, McEwen BF, DeLuca JG (2008) Kinetochore–microtubule attachment relies on the disordered N-terminal tail domain of Hec1. Curr Biol 18:1778–1784. https://doi.org/10.1016/j.cub.2008.08.012
Miller MP, Asbury CL, Biggins S (2016) A TOG protein confers tension sensitivity to kinetochore–microtubule attachments. Cell 165:1428–1439. https://doi.org/10.1016/j.cell.2016.04.030
Pinsky BA, Kung C, Shokat KM, Biggins S (2006) The Ipl1-Aurora protein kinase activates the spindle checkpoint by creating unattached kinetochores. Nat Cell Biol 8:78–83. https://doi.org/10.1038/ncb1341
Liu D, Vleugel M, Backer CB et al (2010) Regulated targeting of protein phosphatase 1 to the outer kinetochore by KNL1 opposes Aurora B kinase. J Cell Biol 188:809–820. https://doi.org/10.1083/jcb.201001006
Emanuele MJ, Lan W, Jwa M et al (2008) Aurora B kinase and protein phosphatase 1 have opposing roles in modulating kinetochore assembly. J Cell Biol 181:241–254. https://doi.org/10.1083/jcb.200710019
Francisco L, Wang W, Chan CS (1994) Type 1 protein phosphatase acts in opposition to IpL1 protein kinase in regulating yeast chromosome segregation. Mol Cell Biol 14:4731–4740. https://doi.org/10.1128/mcb.14.7.4731
Hsu JY, Sun ZW, Li X et al (2000) Mitotic phosphorylation of histone H3 is governed by Ipl1/aurora kinase and Glc7/PP1 phosphatase in budding yeast and nematodes. Cell 102:279–291. https://doi.org/10.1016/s0092-8674(00)00034-9
Vanoosthuyse V, Hardwick KG (2009) A novel protein phosphatase 1-dependent spindle checkpoint silencing mechanism. Curr Biol 19:1176–1181. https://doi.org/10.1016/j.cub.2009.05.060
Etemad B, Kuijt TEF, Kops GJPL (2015) Kinetochore–microtubule attachment is sufficient to satisfy the human spindle assembly checkpoint. Nat Commun 6:8987–8988. https://doi.org/10.1038/ncomms9987
Tauchman EC, Boehm FJ, DeLuca JG (2015) Stable kinetochore–microtubule attachment is sufficient to silence the spindle assembly checkpoint in human cells. Nat Commun 6:10036–10039. https://doi.org/10.1038/ncomms10036
Hori T, Amano M, Suzuki A et al (2008) CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell 135:1039–1052. https://doi.org/10.1016/j.cell.2008.10.019
Nishino T, Takeuchi K, Gascoigne KE et al (2012) CENP-T-W-S-X forms a unique centromeric chromatin structure with a histone-like fold. Cell 148:487–501. https://doi.org/10.1016/j.cell.2011.11.061
Carroll CW, Silva MCC, Godek KM et al (2009) Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat Cell Biol 11:896–902. https://doi.org/10.1038/ncb1899
Carroll CW, Milks KJ, Straight AF (2010) Dual recognition of CENP-A nucleosomes is required for centromere assembly. J Cell Biol 189:1143–1155. https://doi.org/10.1083/jcb.201001013
McKinley KL, Sekulic N, Guo LY et al (2015) The CENP-L-N complex forms a critical node in an integrated meshwork of interactions at the centromere-kinetochore interface. Mol Cell 60:886–898. https://doi.org/10.1016/j.molcel.2015.10.027
Kato H, Jiang J, Zhou B-R et al (2013) A conserved mechanism for centromeric nucleosome recognition by centromere protein CENP-C. Science 340:1110–1113. https://doi.org/10.1126/science.1235532
Pentakota S, Zhou K, Smith C et al (2017) Decoding the centromeric nucleosome through CENP-N. Elife 6:213. https://doi.org/10.7554/eLife.33442
Chittori S, Hong J, Saunders H et al (2018) Structural mechanisms of centromeric nucleosome recognition by the kinetochore protein CENP-N. Science 359:339–343. https://doi.org/10.1126/science.aar2781
Tian T, Li X, Liu Y et al (2018) Molecular basis for CENP-N recognition of CENP-A nucleosome on the human kinetochore. Cell Res 28:374–378. https://doi.org/10.1038/cr.2018.13
Allu PK, Dawicki-McKenna JM, Van Eeuwen T et al (2019) Structure of the human core centromeric nucleosome complex. Curr Biol 29:2625–2639.e5. https://doi.org/10.1016/j.cub.2019.06.062
Tachiwana H, Kagawa W, Shiga T et al (2011) Crystal structure of the human centromeric nucleosome containing CENP-A. Nature 476:232–235. https://doi.org/10.1038/nature10258
Guo LY, Allu PK, Zandarashvili L et al (2017) Centromeres are maintained by fastening CENP-A to DNA and directing an arginine anchor-dependent nucleosome transition. Nat Commun 8:15775. https://doi.org/10.1038/ncomms15775
Nagpal H, Hori T, Furukawa A et al (2015) Dynamic changes in CCAN organization through CENP-C during cell-cycle progression. Mol Biol Cell 26:3768–3776. https://doi.org/10.1091/mbc.E15-07-0531
Fukagawa T, Mikami Y, Nishihashi A et al (2001) CENP-H, a constitutive centromere component, is required for centromere targeting of CENP-C in vertebrate cells. EMBO J 20:4603–4617. https://doi.org/10.1093/emboj/20.16.4603
Kwon M-S, Hori T, Okada M, Fukagawa T (2007) CENP-C is involved in chromosome segregation, mitotic checkpoint function, and kinetochore assembly. Mol Biol Cell 18:2155–2168. https://doi.org/10.1091/mbc.E07-01-0045
Fang J, Liu Y, Wei Y et al (2015) Structural transitions of centromeric chromatin regulate the cell cycle-dependent recruitment of CENP-N. Genes Dev 29:1058–1073. https://doi.org/10.1101/gad.259432.115
Hellwig D, Emmerth S, Ulbricht T et al (2011) Dynamics of CENP-N kinetochore binding during the cell cycle. J Cell Sci 124:3871–3883. https://doi.org/10.1242/jcs.088625
Kline SL, Cheeseman IM, Hori T et al (2006) The human Mis12 complex is required for kinetochore assembly and proper chromosome segregation. J Cell Biol 173:9–17. https://doi.org/10.1083/jcb.200509158
Nishino T, Rago F, Hori T et al (2013) CENP-T provides a structural platform for outer kinetochore assembly. EMBO J 32:424–436. https://doi.org/10.1038/emboj.2012.348
Takeuchi K, Nishino T, Mayanagi K et al (2014) The centromeric nucleosome-like CENP-T-W-S-X complex induces positive supercoils into DNA. Nucleic Acids Res 42:1644–1655. https://doi.org/10.1093/nar/gkt1124
Hara M, Fukagawa T (2019) Where is the right path heading from the centromere to spindle microtubules? Cell Cycle 18:1199–1211. https://doi.org/10.1080/15384101.2019.1617008
Hori T, Shang W-H, Takeuchi K, Fukagawa T (2013) The CCAN recruits CENP-A to the centromere and forms the structural core for kinetochore assembly. J Cell Biol 200:45–60. https://doi.org/10.1083/jcb.201210106
Drinnenberg IA, Akiyoshi B (2017) Evolutionary lessons from species with unique kinetochores. Prog Mol Subcell Biol 56:111–138. https://doi.org/10.1007/978-3-319-58592-5_5
Drinnenberg IA, Henikoff S, Malik HS (2016) Evolutionary turnover of kinetochore proteins: a ship of theseus? Trends Cell Biol 26:498–510. https://doi.org/10.1016/j.tcb.2016.01.005
van Hooff JJ, Tromer E, van Wijk LM et al (2017) Evolutionary dynamics of the kinetochore network in eukaryotes as revealed by comparative genomics. EMBO Rep 18:1559–1571. https://doi.org/10.15252/embr.201744102
Gascoigne KE, Takeuchi K, Suzuki A et al (2011) Induced ectopic kinetochore assembly bypasses the requirement for CENP-A nucleosomes. Cell 145:410–422. https://doi.org/10.1016/j.cell.2011.03.031
Przewloka MR, Venkei Z, Bolanos-Garcia VM et al (2011) CENP-C is a structural platform for kinetochore assembly. Curr Biol 21:399–405. https://doi.org/10.1016/j.cub.2011.02.005
Screpanti E, De Antoni A, Alushin GM et al (2011) Direct binding of Cenp-C to the Mis12 complex joins the inner and outer kinetochore. Curr Biol 21:391–398. https://doi.org/10.1016/j.cub.2010.12.039
Petrovic A, Keller J, Liu Y et al (2016) Structure of the MIS12 complex and molecular basis of its interaction with CENP-C at human kinetochores. Cell 167:1028–1040.e15. https://doi.org/10.1016/j.cell.2016.10.005
Kim S, Yu H (2015) Multiple assembly mechanisms anchor the KMN spindle checkpoint platform at human mitotic kinetochores. J Cell Biol 208:181–196. https://doi.org/10.1083/jcb.201407074
Dimitrova YN, Jenni S, Valverde R et al (2016) Structure of the MIND complex defines a regulatory focus for yeast kinetochore assembly. Cell 167:1014–1027.e12. https://doi.org/10.1016/j.cell.2016.10.011
Rago F, Gascoigne KE, Cheeseman IM (2015) Distinct organization and regulation of the outer kinetochore KMN network downstream of CENP-C and CENP-T. Curr Biol 25:671–677. https://doi.org/10.1016/j.cub.2015.01.059
Huis In't Veld PJ, Jeganathan S, Petrovic A et al (2016) Molecular basis of outer kinetochore assembly on CENP-T. Elife. https://doi.org/10.7554/eLife.21007
Malvezzi F, Litos G, Schleiffer A et al (2013) A structural basis for kinetochore recruitment of the Ndc80 complex via two distinct centromere receptors. EMBO J 32:409–423. https://doi.org/10.1038/emboj.2012.356
Vigneron S, Robert P, Hached K et al (2016) The master Greatwall kinase, a critical regulator of mitosis and meiosis. Int J Dev Biol 60:245–254. https://doi.org/10.1387/ijdb.160155tl
Suzuki A, Badger BL, Wan X et al (2014) The architecture of CCAN proteins creates a structural integrity to resist spindle forces and achieve proper Intrakinetochore stretch. Dev Cell 30:717–730. https://doi.org/10.1016/j.devcel.2014.08.003
Acknowledgements
We thank all members in the Fukagawa lab for helpful discussions. This work was supported by JSPS KAKENHI Grant Numbers 15H05972, 16H06279, and 17H06167 to TF, JSPS KAKENHI Grant Number 16K18491 to MH.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Hara, M., Fukagawa, T. Dynamics of kinetochore structure and its regulations during mitotic progression. Cell. Mol. Life Sci. 77, 2981–2995 (2020). https://doi.org/10.1007/s00018-020-03472-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00018-020-03472-4