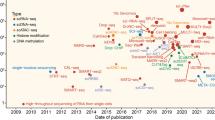
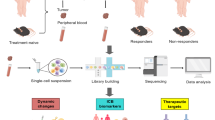
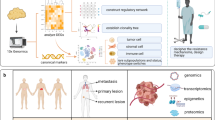
Abstract
Recently, immunotherapy has gained increasing popularity in oncology. Several immunotherapies obtained remarkable clinical effects, but the efficacy varied, and only subsets of cancer patients benefited. Breaking the constraints and improving immunotherapy efficacy is extremely important in precision medicine. Whereas traditional sequencing approaches mask the characteristics of individual cells, single-cell sequencing provides multiple dimensions of cellular characterization at the single-cell level, including genomic, transcriptomic, epigenomic, proteomic, and multi-omics. Hence, the complexity of the tumor microenvironment, the universality of tumor heterogeneity, cell composition and cell–cell interactions, cell lineage tracking, and tumor drug resistance mechanisms are revealed in-depth. However, the clinical transformation of single-cell technology is not to the point of in-depth study, especially in the application of immunotherapy. The newly discovered vital cells and tremendous biomarkers facilitate the development of more efficient individualized therapeutic regimens to guide clinical treatment and predict prognosis. This review provided an overview of the progress in distinct single-cell sequencing methods and emerging strategies. For perspective, the expanding utility of combining single-cell sequencing and other technologies was discussed.



Similar content being viewed by others
Data availability
Not applicable.
Abbreviations
- IFN-α:
-
Interferon-alpha
- ICIs:
-
Immune checkpoint inhibitors
- ACT:
-
Adoptive cellular immunotherapy
- FDA:
-
United States food and drug administration
- CAR-T:
-
Chimeric antigen receptor-T cell
- CAR-NK:
-
Chimeric antigen receptor-engineered natural killer cell
- HCC:
-
Hepatocellular carcinoma
- CAR-M:
-
Chimeric antigen receptor macrophage
- TME:
-
Tumor microenvironment
- TMB:
-
Tumor mutational burden
- ITH:
-
Intra-tumoral heterogeneity
- scRNA-seq:
-
Single-cell RNA sequencing
- FACS:
-
Fluorescence-activated cell sorting
- MACS:
-
Magnetic-activated cell sorting
- UMIs:
-
Unique molecular identifiers
- LCM:
-
Laser capture microdissection
- snRNA-seq:
-
Single-nucleus RNA sequencing
- cDNAs:
-
Complementary DNAs
- PCR:
-
Polymerase chain reaction
- IVT:
-
In vitro transcription
- TCR:
-
T cell receptor
- MHC:
-
Major histocompatibility complex
- PDAC:
-
Pancreatic ductal adenocarcinoma
- scDNA-seq:
-
Single-cell DNA sequencing
- WGS:
-
Whole-genome sequencing
- WES:
-
Whole-exome sequencing
- SNVs:
-
Single nucleotide variants
- CNAs:
-
Copy number aberrations
- WGBS:
-
Whole-genome bisulfite sequencing
- CyTOF:
-
Cyclic immunofluorescence
- CNVs:
-
Copy number variations
- TIME:
-
Tumor immune microenvironment
- CRC:
-
Colorectal cancer
- NSCLC:
-
Non-small cell lung cancer
- CTCs:
-
Circulating tumor cells
- CSCs:
-
Cancer stem cells
- SCLC:
-
Small-cell lung cancer
- IMC:
-
Immature myeloid cell
- TNF:
-
Tumor necrosis factor
- iCD:
-
Ileal crohn’s disease
- RCC:
-
Renal cell carcinoma
- cHL:
-
Classical hodgkin lymphoma
- GVAX:
-
Granulocyte–macrophage colony-stimulating factor (GM-CSF)-allogeneic pancreatic tumor cells
- OS:
-
Overall survival
- CyCIF:
-
Cyclic immunofluorescence
- MIBI:
-
Multiplexed ion beam imaging
- cSCC:
-
Cutaneous squamous cell carcinoma
References
Yang F, Shi K, Jia Y-P, Hao Y, Peng J-R, Qian Z-Y (2020) Advanced biomaterials for cancer immunotherapy. Acta Pharmacol Sin 41(7):911–927. https://doi.org/10.1038/s41401-020-0372-z
Riley RS, June CH, Langer R, Mitchell MJ (2019) Delivery technologies for cancer immunotherapy. Nat Rev Drug Discov 18(3):175–196. https://doi.org/10.1038/s41573-018-0006-z
Hu W, Huang F, Ning L, Hao J, Wan J, Hao S (2020) Enhanced immunogenicity of leukemia-derived exosomes via transfection with lentiviral vectors encoding costimulatory molecules. Cell Oncol (Dordr) 43(5):889–900. https://doi.org/10.1007/s13402-020-00535-3
Carrega P, Loiacono F, Di Carlo E, Scaramuccia A, Mora M, Conte R et al (2015) NCR(+)ILC3 concentrate in human lung cancer and associate with intratumoral lymphoid structures. Nat Commun. https://doi.org/10.1038/ncomms9280
Qin S, Xu L, Yi M, Yu S, Wu K, Luo S (2019) Novel immune checkpoint targets: moving beyond PD-1 and CTLA-4. Mol Cancer 18(1):155. https://doi.org/10.1186/s12943-019-1091-2
June CH, O’Connor RS, Kawalekar OU, Ghassemi S, Milone MC (2018) CAR T cell immunotherapy for human cancer. Science 359(6382):1361–1365. https://doi.org/10.1126/science.aar6711
Ma R, Lu T, Li Z, Teng K-Y, Mansour AG, Yu M et al (2021) An oncolytic virus expressing IL15/IL15Rα combined with off-the-shelf EGFR-CAR NK cells targets glioblastoma. Cancer Res 81(13):3635–3648. https://doi.org/10.1158/0008-5472.CAN-21-0035
Li Y, Hermanson DL, Moriarity BS, Kaufman DS (2018) Human iPSC-derived natural killer cells engineered with chimeric antigen receptors enhance anti-tumor activity. Cell Stem Cell. https://doi.org/10.1016/j.stem.2018.06.002
Mantovani S, Oliviero B, Varchetta S, Mele D, Mondelli MU (2020) Natural killer cell responses in hepatocellular carcinoma: implications for novel immunotherapeutic approaches. Cancers. https://doi.org/10.3390/cancers12040926
Cotechini T, Atallah A, Grossman A (2021) Tissue-resident and recruited macrophages in primary tumor and metastatic microenvironments: potential targets in cancer therapy. Cells. https://doi.org/10.3390/cells10040960
Sklavenitis-Pistofidis R, Getz G, Ghobrial I (2021) Single-cell RNA sequencing: one step closer to the clinic. Nat Med 27(3):375–376. https://doi.org/10.1038/s41591-021-01276-y
Kas B, Talbot H, Ferrara R, Richard C, Lamarque JP, Pitre-Champagnat S et al (2020) Clarification of definitions of hyperprogressive disease during immunotherapy for non-small cell lung cancer. JAMA Oncol 6(7):1039–1046. https://doi.org/10.1001/jamaoncol.2020.1634
Pilanc P, Wojnicki K, Roura A-J, Cyranowski S, Ellert-Miklaszewska A, Ochocka N et al (2021) A novel oral arginase 1/2 inhibitor enhances the antitumor effect of PD-1 inhibition in murine experimental gliomas by altering the immunosuppressive environment. Front Oncol. https://doi.org/10.3389/fonc.2021.703465
Luke JJ, Lemons JM, Karrison TG, Pitroda SP, Melotek JM, Zha Y et al (2018) Safety and clinical activity of pembrolizumab and multisite stereotactic body radiotherapy in patients with advanced solid tumors. J Clin Oncol: Off J Am Soc Clin Oncol 36(16):1611–1618. https://doi.org/10.1200/JCO.2017.76.2229
Aurisicchio L, Pallocca M, Ciliberto G, Palombo F (2018) The perfect personalized cancer therapy: cancer vaccines against neoantigens. J Exp Clin Cancer Res 37(1):86. https://doi.org/10.1186/s13046-018-0751-1
Potter SS (2018) Single-cell RNA sequencing for the study of development, physiology and disease. Nat Rev Nephrol 14(8):479–492. https://doi.org/10.1038/s41581-018-0021-7
Zhou Y, Bian S, Zhou X, Cui Y, Wang W, Wen L et al (2020) Single-cell multiomics sequencing reveals prevalent genomic alterations in tumor stromal cells of human colorectal cancer. Cancer Cell. https://doi.org/10.1016/j.ccell.2020.09.015
Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N et al (2009) mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods 6(5):377–382. https://doi.org/10.1038/nmeth.1315
Hedlund E, Deng Q (2018) Single-cell RNA sequencing: technical advancements and biological applications. Mol Aspects Med 59:36–46. https://doi.org/10.1016/j.mam.2017.07.003
Reichard A, Asosingh K (2019) Best practices for preparing a single cell suspension from solid tissues for flow cytometry. Cytometry Part A: J Intl Soc Analytical Cytol 95(2):219–226. https://doi.org/10.1002/cyto.a.23690
Hwang H-W, Saito Y, Park CY, Blachère NE, Tajima Y, Fak JJ et al (2017) cTag-PAPERCLIP reveals alternative polyadenylation promotes cell-type specific protein diversity and shifts araf isoforms with microglia activation. Neuron. https://doi.org/10.1016/j.neuron.2017.08.024
Gross A, Schoendube J, Zimmermann S, Steeb M, Zengerle R, Koltay P (2015) Technologies for single-cell isolation. Int J Mol Sci 16(8):16897–16919. https://doi.org/10.3390/ijms160816897
Shen M-J, Olsthoorn RCL, Zeng Y, Bakkum T, Kros A, Boyle AL (2021) Magnetic-activated cell sorting using coiled-coil peptides: an alternative strategy for isolating cells with high efficiency and specificity. ACS Appl Mater Interfaces 13(10):11621–11630. https://doi.org/10.1021/acsami.0c22185
Xin Y, Kim J, Ni M, Wei Y, Okamoto H, Lee J et al (2016) Use of the Fluidigm C1 platform for RNA sequencing of single mouse pancreatic islet cells. Proc Natl Acad Sci USA 113(12):3293–3298. https://doi.org/10.1073/pnas.1602306113
Briggs JA, Weinreb C, Wagner DE, Megason S, Peshkin L, Kirschner MW et al (2018) The dynamics of gene expression in vertebrate embryogenesis at single-cell resolution. Science. https://doi.org/10.1126/science.aar5780
Farrell JA, Wang Y, Riesenfeld SJ, Shekhar K, Regev A, Schier AF (2018) Single-cell reconstruction of developmental trajectories during zebrafish embryogenesis. Science. https://doi.org/10.1126/science.aar3131
Zheng GXY, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R et al (2017) Massively parallel digital transcriptional profiling of single cells. Nat Commun. https://doi.org/10.1038/ncomms14049
Romagnoli D, Boccalini G, Bonechi M, Biagioni C, Fassan P, Bertorelli R et al (2018) ddSeeker: a tool for processing Bio-Rad ddSEQ single cell RNA-seq data. BMC Genom 19(1):960. https://doi.org/10.1186/s12864-018-5249-x
Ding J, Adiconis X, Simmons SK, Kowalczyk MS, Hession CC, Marjanovic ND et al (2020) Systematic comparison of single-cell and single-nucleus RNA-sequencing methods. Nat Biotechnol 38(6):737–746. https://doi.org/10.1038/s41587-020-0465-8
Zhang X, Li T, Liu F, Chen Y, Yao J, Li Z et al (2019) Comparative analysis of droplet-based ultra-high-throughput single-cell RNA-seq systems. Mol Cell. https://doi.org/10.1016/j.molcel.2018.10.020
Foley JW, Zhu C, Jolivet P, Zhu SX, Lu P, Meaney MJ et al (2019) Gene expression profiling of single cells from archival tissue with laser-capture microdissection and smart-3SEQ. Genome Res 29(11):1816–1825. https://doi.org/10.1101/gr.234807.118
Schellenberg A, Stiehl T, Horn P, Joussen S, Pallua N, Ho AD et al (2012) Population dynamics of mesenchymal stromal cells during culture expansion. Cytotherapy 14(4):401–411. https://doi.org/10.3109/14653249.2011.640669
Habib N, Avraham-Davidi I, Basu A, Burks T, Shekhar K, Hofree M et al (2017) Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat Methods 14(10):955–958. https://doi.org/10.1038/nmeth.4407
Lei Y, Tang R, Xu J, Wang W, Zhang B, Liu J et al (2021) Applications of single-cell sequencing in cancer research: progress and perspectives. J Hematol Oncol 14(1):91. https://doi.org/10.1186/s13045-021-01105-2
Slyper M, Porter CBM, Ashenberg O, Waldman J, Drokhlyansky E, Wakiro I et al (2020) A single-cell and single-nucleus RNA-Seq toolbox for fresh and frozen human tumors. Nat Med 26(5):792–802. https://doi.org/10.1038/s41591-020-0844-1
Jovic D, Liang X, Zeng H, Lin L, Xu F, Luo Y (2022) Single-cell RNA sequencing technologies and applications: a brief overview. Clin Transl Med 12(3):e694. https://doi.org/10.1002/ctm2.694
Gibellini L, De Biasi S, Porta C, Lo Tartaro D, Depenni R, Pellacani G et al (2020) Single-cell approaches to profile the response to immune checkpoint inhibitors. Front Immunol. https://doi.org/10.3389/fimmu.2020.00490
Li B, Li T, Pignon J-C, Wang B, Wang J, Shukla SA et al (2016) Landscape of tumor-infiltrating T cell repertoire of human cancers. Nat Genet 48(7):725–732. https://doi.org/10.1038/ng.3581
Bradley P, Thomas PG (2019) Using T cell receptor repertoires to understand the principles of adaptive immune recognition. Annu Rev Immunol 37:547–570. https://doi.org/10.1146/annurev-immunol-042718-041757
Zheng C, Zheng L, Yoo J-K, Guo H, Zhang Y, Guo X et al (2017) Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell. https://doi.org/10.1016/j.cell.2017.05.035
Ye L, Creaney J, Redwood A, Robinson B (2021) The current lung cancer neoantigen landscape and implications for therapy. J Thoracic Oncol: Off Publ Int Assoc Study Lung Cancer 16(6):922–932. https://doi.org/10.1016/j.jtho.2021.01.1624
Rosenthal R, Cadieux EL, Salgado R, Bakir MA, Moore DA, Hiley CT et al (2019) Neoantigen-directed immune escape in lung cancer evolution. Nature 567(7749):479–485. https://doi.org/10.1038/s41586-019-1032-7
Blass E, Ott PA (2021) Advances in the development of personalized neoantigen-based therapeutic cancer vaccines. Nat Rev Clin Oncol 18(4):215–229. https://doi.org/10.1038/s41571-020-00460-2
Balachandran VP, Łuksza M, Zhao JN, Makarov V, Moral JA, Remark R et al (2017) Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature 551(7681):512–516. https://doi.org/10.1038/nature24462
Cafri G, Gartner JJ, Zaks T, Hopson K, Levin N, Paria BC et al (2020) mRNA vaccine-induced neoantigen-specific T cell immunity in patients with gastrointestinal cancer. J Clin Investig 130(11):5976–5988. https://doi.org/10.1172/JCI134915
Rodriques SG, Stickels RR, Goeva A, Martin CA, Murray E, Vanderburg CR et al (2019) Slide-seq: a scalable technology for measuring genome-wide expression at high spatial resolution. Science 363(6434):1463–1467. https://doi.org/10.1126/science.aaw1219
Buenrostro JD, Wu B, Litzenburger UM, Ruff D, Gonzales ML, Snyder MP et al (2015) Single-cell chromatin accessibility reveals principles of regulatory variation. Nature 523(7561):486–490. https://doi.org/10.1038/nature14590
Gawad C, Koh W, Quake SR (2016) Single-cell genome sequencing: current state of the science. Nat Rev Genet 17(3):175–188. https://doi.org/10.1038/nrg.2015.16
Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J et al (2011) Tumour evolution inferred by single-cell sequencing. Nature 472(7341):90–94. https://doi.org/10.1038/nature09807
Bakker B, Taudt A, Belderbos ME, Porubsky D, Spierings DCJ, de Jong TV et al (2016) Single-cell sequencing reveals karyotype heterogeneity in murine and human malignancies. Genome Biol 17(1):115. https://doi.org/10.1186/s13059-016-0971-7
Baslan T, Kendall J, Volyanskyy K, McNamara K, Cox H, D’Italia S et al (2020) Novel insights into breast cancer copy number genetic heterogeneity revealed by single-cell genome sequencing. Elife. https://doi.org/10.7554/eLife.51480
Ferronika P, van den Bos H, Taudt A, Spierings DCJ, Saber A, Hiltermann TJN et al (2017) Copy number alterations assessed at the single-cell level revealed mono- and polyclonal seeding patterns of distant metastasis in a small-cell lung cancer patient. Ann Oncol 28(7):1668–1670. https://doi.org/10.1093/annonc/mdx182
Xu X, Hou Y, Yin X, Bao L, Tang A, Song L et al (2012) Single-cell exome sequencing reveals single-nucleotide mutation characteristics of a kidney tumor. Cell 148(5):886–895. https://doi.org/10.1016/j.cell.2012.02.025
Francis JM, Zhang C-Z, Maire CL, Jung J, Manzo VE, Adalsteinsson VA et al (2014) EGFR variant heterogeneity in glioblastoma resolved through single-nucleus sequencing. Cancer Discov 4(8):956–971. https://doi.org/10.1158/2159-8290.CD-13-0879
Xu W, Wen Y, Liang Y, Xu Q, Wang X, Jin W et al (2021) A plate-based single-cell ATAC-seq workflow for fast and robust profiling of chromatin accessibility. Nat Protoc 16(8):4084–4107. https://doi.org/10.1038/s41596-021-00583-5
Muto Y, Wilson PC, Ledru N, Wu H, Dimke H, Waikar SS et al (2021) Single cell transcriptional and chromatin accessibility profiling redefine cellular heterogeneity in the adult human kidney. Nat Commun 12(1):2190. https://doi.org/10.1038/s41467-021-22368-w
Cokus SJ, Feng S, Zhang X, Chen Z, Merriman B, Haudenschild CD et al (2008) Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452(7184):215–219. https://doi.org/10.1038/nature06745
Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A et al (2008) Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 454(7205):766–770. https://doi.org/10.1038/nature07107
Chen Z, Chen JJ, Fan R (2019) Single-cell protein secretion detection and profiling. Annu Rev Anal Chem (Palo Alto, Calif) 12(1):431–449. https://doi.org/10.1146/annurev-anchem-061318-115055
Petelski AA, Emmott E, Leduc A, Huffman RG, Specht H, Perlman DH et al (2021) Multiplexed single-cell proteomics using SCoPE2. Nat Protoc 16(12):5398–5425. https://doi.org/10.1038/s41596-021-00616-z
Budnik B, Levy E, Harmange G, Slavov N (2018) SCoPE-MS: mass spectrometry of single mammalian cells quantifies proteome heterogeneity during cell differentiation. Genome Biol 19(1):161. https://doi.org/10.1186/s13059-018-1547-5
Specht H, Emmott E, Petelski AA, Huffman RG, Perlman DH, Serra M et al (2021) Single-cell proteomic and transcriptomic analysis of macrophage heterogeneity using SCoPE2. Genome Biol 22(1):50. https://doi.org/10.1186/s13059-021-02267-5
Palit S, Heuser C, de Almeida GP, Theis FJ, Zielinski CE (2019) Meeting the challenges of high-dimensional single-cell data analysis in immunology. Front Immunol. https://doi.org/10.3389/fimmu.2019.01515
Gilmore IS, Heiles S, Pieterse CL (2019) Metabolic imaging at the single-cell scale: recent advances in mass spectrometry imaging. Annu Rev Anal Chem (Palo Alto, Calif) 12(1):201–224. https://doi.org/10.1146/annurev-anchem-061318-115516
Fanucchi S, Domínguez-Andrés J, Joosten LAB, Netea MG, Mhlanga MM (2021) The intersection of epigenetics and metabolism in trained immunity. Immunity 54(1):32–43. https://doi.org/10.1016/j.immuni.2020.10.011
Artyomov MN, Van den Bossche J (2020) Immunometabolism in the single-cell era. Cell Metab 32(5):710–725. https://doi.org/10.1016/j.cmet.2020.09.013
Van den Bossche J, O’Neill LA, Menon D (2017) Macrophage immunometabolism: where are we (going)? Trends Immunol 38(6):395–406. https://doi.org/10.1016/j.it.2017.03.001
Mazumdar C, Driggers EM, Turka LA (2020) The Untapped opportunity and challenge of immunometabolism: a new paradigm for drug discovery. Cell Metab 31(1):26–34. https://doi.org/10.1016/j.cmet.2019.11.014
Nam AS, Chaligne R, Landau DA (2021) Integrating genetic and non-genetic determinants of cancer evolution by single-cell multi-omics. Nat Rev Genet. https://doi.org/10.1038/s41576-020-0265-5
Rodriguez-Meira A, Buck G, Clark S-A, Povinelli BJ, Alcolea V, Louka E et al (2019) Unravelling intratumoral heterogeneity through high-sensitivity single-cell mutational analysis and parallel RNA sequencing. Mol Cell. https://doi.org/10.1016/j.molcel.2019.01.009
Chen S, Lake BB, Zhang K (2019) High-throughput sequencing of the transcriptome and chromatin accessibility in the same cell. Nat Biotechnol 37(12):1452–1457. https://doi.org/10.1038/s41587-019-0290-0
Stoeckius M, Hafemeister C, Stephenson W, Houck-Loomis B, Chattopadhyay PK, Swerdlow H et al (2017) Simultaneous epitope and transcriptome measurement in single cells. Nat Methods 14(9):865–868. https://doi.org/10.1038/nmeth.4380
Cao J, Cusanovich DA, Ramani V, Aghamirzaie D, Pliner HA, Hill AJ et al (2018) Joint profiling of chromatin accessibility and gene expression in thousands of single cells. Science 361(6409):1380–1385. https://doi.org/10.1126/science.aau0730
Argelaguet R, Velten B, Arnol D, Dietrich S, Zenz T, Marioni JC et al (2018) Multi-omics factor analysis-a framework for unsupervised integration of multi-omics data sets. Mol Syst Biol 14(6):e8124. https://doi.org/10.15252/msb.20178124
Mair F, Erickson JR, Voillet V, Simoni Y, Bi T, Tyznik AJ et al (2020) A targeted multi-omic analysis approach measures protein expression and low-abundance transcripts on the single-cell level. Cell Rep 31(1):107499. https://doi.org/10.1016/j.celrep.2020.03.063
Vitak SA, Torkenczy KA, Rosenkrantz JL, Fields AJ, Christiansen L, Wong MH et al (2017) Sequencing thousands of single-cell genomes with combinatorial indexing. Nat Methods 14(3):302–308. https://doi.org/10.1038/nmeth.4154
Eyler CE, Matsunaga H, Hovestadt V, Vantine SJ, van Galen P, Bernstein BE (2020) Single-cell lineage analysis reveals genetic and epigenetic interplay in glioblastoma drug resistance. Genome Biol 21(1):174. https://doi.org/10.1186/s13059-020-02085-1
Casasent AK, Schalck A, Gao R, Sei E, Long A, Pangburn W et al (2018) Multiclonal invasion in breast tumors identified by topographic single cell sequencing. Cell. https://doi.org/10.1016/j.cell.2017.12.007
Leung ML, Davis A, Gao R, Casasent A, Wang Y, Sei E et al (2017) Single-cell DNA sequencing reveals a late-dissemination model in metastatic colorectal cancer. Genome Res 27(8):1287–1299. https://doi.org/10.1101/gr.209973.116
Mooijman D, Dey SS, Boisset J-C, Crosetto N, van Oudenaarden A (2016) Single-cell 5hmC sequencing reveals chromosome-wide cell-to-cell variability and enables lineage reconstruction. Nat Biotechnol 34(8):852–856. https://doi.org/10.1038/nbt.3598
Raj B, Wagner DE, McKenna A, Pandey S, Klein AM, Shendure J et al (2018) Simultaneous single-cell profiling of lineages and cell types in the vertebrate brain. Nat Biotechnol 36(5):442–450. https://doi.org/10.1038/nbt.4103
Kester L, van Oudenaarden A (2018) Single-cell transcriptomics meets lineage tracing. Cell Stem Cell 23(2):166–179. https://doi.org/10.1016/j.stem.2018.04.014
Zhu X, Wolfgruber TK, Tasato A, Arisdakessian C, Garmire DG, Garmire LX (2017) Granatum: a graphical single-cell RNA-Seq analysis pipeline for genomics scientists. Genome Med 9(1):108. https://doi.org/10.1186/s13073-017-0492-3
Liu Z, Lou H, Xie K, Wang H, Chen N, Aparicio OM et al (2017) Reconstructing cell cycle pseudo time-series via single-cell transcriptome data. Nat Commun 8(1):22. https://doi.org/10.1038/s41467-017-00039-z
La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V et al (2018) RNA velocity of single cells. Nature 560(7719):494–498. https://doi.org/10.1038/s41586-018-0414-6
Bergen V, Lange M, Peidli S, Wolf FA, Theis FJ (2020) Generalizing RNA velocity to transient cell states through dynamical modeling. Nat Biotechnol 38(12):1408–1414. https://doi.org/10.1038/s41587-020-0591-3
Qiu X, Zhang Y, Martin-Rufino JD, Weng C, Hosseinzadeh S, Yang D et al (2022) Mapping transcriptomic vector fields of single cells. Cell. https://doi.org/10.1016/j.cell.2021.12.045
Zhou L, Yu KH, Wong TL, Zhang Z, Chan CH, Loong JH et al (2021) Lineage tracing and single-cell analysis reveal proliferative Prom1+ tumour-propagating cells and their dynamic cellular transition during liver cancer progression. Gut. https://doi.org/10.1136/gutjnl-2021-324321
Ma L, Wang L, Khatib SA, Chang C-W, Heinrich S, Dominguez DA et al (2021) Single-cell atlas of tumor cell evolution in response to therapy in hepatocellular carcinoma and intrahepatic cholangiocarcinoma. J Hepatol 75(6):1397–1408. https://doi.org/10.1016/j.jhep.2021.06.028
Medler TR, Cotechini T, Coussens LM (2015) Immune response to cancer therapy: mounting an effective antitumor response and mechanisms of resistance. Trends In Cancer 1(1):66–75
Geller AE, Shrestha R, Woeste MR, Guo H, Hu X, Ding C et al (2022) The induction of peripheral trained immunity in the pancreas incites anti-tumor activity to control pancreatic cancer progression. Nat Commun 13(1):759. https://doi.org/10.1038/s41467-022-28407-4
Sun L, Yang X, Yuan Z, Wang H (2020) Metabolic reprogramming in immune response and tissue inflammation. Arterioscler Thromb Vasc Biol 40(9):1990–2001. https://doi.org/10.1161/ATVBAHA.120.314037
Yang X, Qi Q, Pan Y, Zhou Q, Wu Y, Zhuang J et al (2020) Single-cell analysis reveals characterization of infiltrating T cells in moderately differentiated colorectal cancer. Front Immunol. https://doi.org/10.3389/fimmu.2020.620196
Chung W, Eum HH, Lee H-O, Lee K-M, Lee H-B, Kim K-T et al (2017) Single-cell RNA-seq enables comprehensive tumour and immune cell profiling in primary breast cancer. Nat Commun. https://doi.org/10.1038/ncomms15081
Wu F, Fan J, He Y, Xiong A, Yu J, Li Y et al (2021) Single-cell profiling of tumor heterogeneity and the microenvironment in advanced non-small cell lung cancer. Nat Commun 12(1):2540. https://doi.org/10.1038/s41467-021-22801-0
Lambrechts D, Wauters E, Boeckx B, Aibar S, Nittner D, Burton O et al (2018) Phenotype molding of stromal cells in the lung tumor microenvironment. Nat Med 24(8):1277–1289. https://doi.org/10.1038/s41591-018-0096-5
Ho DW-H, Tsui Y-M, Sze KM-F, Chan L-K, Cheung T-T, Lee E et al (2019) Single-cell transcriptomics reveals the landscape of intra-tumoral heterogeneity and stemness-related subpopulations in liver cancer. Cancer Lett 459:176–185. https://doi.org/10.1016/j.canlet.2019.06.002
Grosselin K, Durand A, Marsolier J, Poitou A, Marangoni E, Nemati F et al (2019) High-throughput single-cell ChIP-seq identifies heterogeneity of chromatin states in breast cancer. Nat Genet 51(6):1060–1066. https://doi.org/10.1038/s41588-019-0424-9
Zhou Y, Yang D, Yang Q, Lv X, Huang W, Zhou Z et al (2020) Single-cell RNA landscape of intratumoral heterogeneity and immunosuppressive microenvironment in advanced osteosarcoma. Nat Commun 11(1):6322. https://doi.org/10.1038/s41467-020-20059-6
Zhang S, Jiang VC, Han G, Hao D, Lian J, Liu Y et al (2021) Longitudinal single-cell profiling reveals molecular heterogeneity and tumor-immune evolution in refractory mantle cell lymphoma. Nat Commun 12(1):2877. https://doi.org/10.1038/s41467-021-22872-z
Pailler E, Faugeroux V, Oulhen M, Mezquita L, Laporte M, Honoré A et al (2019) Acquired resistance mutations to ALK inhibitors identified by single circulating tumor cell sequencing in -rearranged non-small-cell lung cancer. Clin Cancer Res: Off J Am Assoc Cancer Res 25(22):6671–6682. https://doi.org/10.1158/1078-0432.CCR-19-1176
Grün D, Lyubimova A, Kester L, Wiebrands K, Basak O, Sasaki N et al (2015) Single-cell messenger RNA sequencing reveals rare intestinal cell types. Nature 525(7568):251–255. https://doi.org/10.1038/nature14966
Tirosh I, Venteicher AS, Hebert C, Escalante LE, Patel AP, Yizhak K et al (2016) Single-cell RNA-seq supports a developmental hierarchy in human oligodendroglioma. Nature 539(7628):309–313. https://doi.org/10.1038/nature20123
Torensma R (2018) The dilemma of cure and damage in oligodendroglioma: ways to tip the balance away from the damage. Cancers. https://doi.org/10.3390/cancers10110431
de Kanter JK, Lijnzaad P, Candelli T, Margaritis T, Holstege FCP (2019) CHETAH: a selective, hierarchical cell type identification method for single-cell RNA sequencing. Nucleic Acids Res 47(16):e95. https://doi.org/10.1093/nar/gkz543
Iorgulescu JB, Braun D, Oliveira G, Keskin DB, Wu CJ (2018) Acquired mechanisms of immune escape in cancer following immunotherapy. Genome Med 10(1):87. https://doi.org/10.1186/s13073-018-0598-2
Chen Y-P, Yin J-H, Li W-F, Li H-J, Chen D-P, Zhang C-J et al (2020) Single-cell transcriptomics reveals regulators underlying immune cell diversity and immune subtypes associated with prognosis in nasopharyngeal carcinoma. Cell Res 30(11):1024–1042. https://doi.org/10.1038/s41422-020-0374-x
Mahata B, Zhang X, Kolodziejczyk AA, Proserpio V, Haim-Vilmovsky L, Taylor AE et al (2014) Single-cell RNA sequencing reveals T helper cells synthesizing steroids de novo to contribute to immune homeostasis. Cell Rep 7(4):1130–1142. https://doi.org/10.1016/j.celrep.2014.04.011
Zhang Q, Rong Y, Yi K, Huang L, Chen M, Wang F (2020) Circulating tumor cells in hepatocellular carcinoma: single-cell based analysis, preclinical models, and clinical applications. Theranostics 10(26):12060–12071. https://doi.org/10.7150/thno.48918
Gkountela S, Castro-Giner F, Szczerba BM, Vetter M, Landin J, Scherrer R et al (2019) Circulating tumor cell clustering shapes DNA methylation to enable metastasis seeding. Cell. https://doi.org/10.1016/j.cell.2018.11.046
Aceto N, Bardia A, Miyamoto DT, Donaldson MC, Wittner BS, Spencer JA et al (2014) Circulating tumor cell clusters are oligoclonal precursors of breast cancer metastasis. Cell 158(5):1110–1122. https://doi.org/10.1016/j.cell.2014.07.013
Hong X, Roh W, Sullivan RJ, Wong KHK, Wittner BS, Guo H et al (2021) The lipogenic regulator SREBP2 induces transferrin in circulating melanoma cells and suppresses ferroptosis. Cancer Discov 11(3):678–695. https://doi.org/10.1158/2159-8290.CD-19-1500
Povinelli BJ, Rodriguez-Meira A, Mead AJ (2018) Single cell analysis of normal and leukemic hematopoiesis. Mol Aspects Med 59:85–94. https://doi.org/10.1016/j.mam.2017.08.006
Peng J, Sun B-F, Chen C-Y, Zhou J-Y, Chen Y-S, Chen H et al (2019) Single-cell RNA-seq highlights intra-tumoral heterogeneity and malignant progression in pancreatic ductal adenocarcinoma. Cell Res 29(9):725–738. https://doi.org/10.1038/s41422-019-0195-y
Goel S, DeCristo MJ, Watt AC, BrinJones H, Sceneay J, Li BB et al (2017) CDK4/6 inhibition triggers anti-tumour immunity. Nature 548(7668):471–475. https://doi.org/10.1038/nature23465
Chan JM, Quintanal-Villalonga Á, Gao VR, Xie Y, Allaj V, Chaudhary O et al (2021) Signatures of plasticity, metastasis, and immunosuppression in an atlas of human small cell lung cancer. Cancer Cell. https://doi.org/10.1016/j.ccell.2021.09.008
Dutertre C-A, Becht E, Irac SE, Khalilnezhad A, Narang V, Khalilnezhad S et al (2019) Single-cell analysis of human mononuclear phagocytes reveals subset-defining markers and identifies circulating inflammatory dendritic cells. Immunity. https://doi.org/10.1016/j.immuni.2019.08.008
Sangro B, Melero I, Wadhawan S, Finn RS, Abou-Alfa GK, Cheng A-L et al (2020) Association of inflammatory biomarkers with clinical outcomes in nivolumab-treated patients with advanced hepatocellular carcinoma. J Hepatol 73(6):1460–1469. https://doi.org/10.1016/j.jhep.2020.07.026
Han S, Shuen WH, Wang W-W, Nazim E, Toh HC (2020) Tailoring precision immunotherapy: coming to a clinic soon? ESMO Open. https://doi.org/10.1136/esmoopen-2019-000631
Villanueva J, Vultur A, Herlyn M (2011) Resistance to BRAF inhibitors: unraveling mechanisms and future treatment options. Cancer Res 71(23):7137–7140. https://doi.org/10.1158/0008-5472.CAN-11-1243
Kieffer Y, Hocine HR, Gentric G, Pelon F, Bernard C, Bourachot B et al (2020) Single-cell analysis reveals fibroblast clusters linked to immunotherapy resistance in cancer. Cancer Discov 10(9):1330–1351. https://doi.org/10.1158/2159-8290.CD-19-1384
Dominguez CX, Müller S, Keerthivasan S, Koeppen H, Hung J, Gierke S et al (2020) Single-cell RNA sequencing reveals stromal evolution into LRRC15 myofibroblasts as a determinant of patient response to cancer immunotherapy. Cancer Discov 10(2):232–253. https://doi.org/10.1158/2159-8290.CD-19-0644
Wang Q, Guldner IH, Golomb SM, Sun L, Harris JA, Lu X et al (2019) Single-cell profiling guided combinatorial immunotherapy for fast-evolving CDK4/6 inhibitor-resistant HER2-positive breast cancer. Nat Commun 10(1):3817. https://doi.org/10.1038/s41467-019-11729-1
Bivona TG, Doebele RC (2016) A framework for understanding and targeting residual disease in oncogene-driven solid cancers. Nat Med 22(5):472–478. https://doi.org/10.1038/nm.4091
Jerby-Arnon L, Shah P, Cuoco MS, Rodman C, Su M-J, Melms JC et al (2018) A cancer cell program promotes T cell exclusion and resistance to checkpoint blockade. Cell. https://doi.org/10.1016/j.cell.2018.09.006
Zhang Y, Zhang Z (2020) The history and advances in cancer immunotherapy: understanding the characteristics of tumor-infiltrating immune cells and their therapeutic implications. Cell Mol Immunol 17(8):807–821. https://doi.org/10.1038/s41423-020-0488-6
Jiang Y-Q, Wang Z-X, Zhong M, Shen L-J, Han X, Zou X et al (2021) Investigating mechanisms of response or resistance to immune checkpoint inhibitors by analyzing cell-cell communications in tumors before and after programmed cell death-1 (PD-1) targeted therapy: an integrative analysis using single-cell RNA and Bulk-RNA sequencing data. Oncoimmunology 10(1):1908010. https://doi.org/10.1080/2162402X.2021.1908010
Sehgal K, Portell A, Ivanova EV, Lizotte PH, Mahadevan NR, Greene JR et al (2021) Dynamic single-cell RNA sequencing identifies immunotherapy persister cells following PD-1 blockade. J Clin Investig. https://doi.org/10.1172/JCI135038
Martin JC, Chang C, Boschetti G, Ungaro R, Giri M, Grout JA et al (2019) Single-cell analysis of crohn’s disease lesions identifies a pathogenic cellular module associated with resistance to anti-TNF therapy. Cell. https://doi.org/10.1016/j.cell.2019.08.008
Hiam-Galvez KJ, Allen BM, Spitzer MH (2021) Systemic immunity in cancer. Nat Rev Cancer 21(6):345–359. https://doi.org/10.1038/s41568-021-00347-z
Allen BM, Hiam KJ, Burnett CE, Venida A, DeBarge R, Tenvooren I et al (2020) Systemic dysfunction and plasticity of the immune macroenvironment in cancer models. Nat Med 26(7):1125–1134. https://doi.org/10.1038/s41591-020-0892-6
Yost KE, Satpathy AT, Wells DK, Qi Y, Wang C, Kageyama R et al (2019) Clonal replacement of tumor-specific T cells following PD-1 blockade. Nat Med 25(8):1251–1259. https://doi.org/10.1038/s41591-019-0522-3
Zhang Q, He Y, Luo N, Patel SJ, Han Y, Gao R et al (2019) Landscape and dynamics of single immune cells in hepatocellular carcinoma. Cell. https://doi.org/10.1016/j.cell.2019.10.003
Sun Y, Wu L, Zhong Y, Zhou K, Hou Y, Wang Z et al (2021) Single-cell landscape of the ecosystem in early-relapse hepatocellular carcinoma. Cell. https://doi.org/10.1016/j.cell.2020.11.041
Zhong J, Liu Z, Cai C, Duan X, Deng T, Zeng G (2021) mA modification patterns and tumor immune landscape in clear cell renal carcinoma. J Immunotherapy Cancer. https://doi.org/10.1136/jitc-2020-001646
Ho DW-H, Tsui Y-M, Chan L-K, Sze KM-F, Zhang X, Cheu JW-S et al (2021) Single-cell RNA sequencing shows the immunosuppressive landscape and tumor heterogeneity of HBV-associated hepatocellular carcinoma. Nat Commun 12(1):3684. https://doi.org/10.1038/s41467-021-24010-1
Van den Berge K, Perraudeau F, Soneson C, Love MI, Risso D, Vert J-P et al (2018) Observation weights unlock bulk RNA-seq tools for zero inflation and single-cell applications. Genome Biol 19(1):24. https://doi.org/10.1186/s13059-018-1406-4
Raghavan S, Winter PS, Navia AW, Williams HL, DenAdel A, Lowder KE et al (2021) Microenvironment drives cell state, plasticity, and drug response in pancreatic cancer. Cell. https://doi.org/10.1016/j.cell.2021.11.017
Kumar V, Ramnarayanan K, Sundar R, Padmanabhan N, Srivastava S, Koiwa M et al (2022) Single-cell atlas of lineage states, tumor microenvironment, and subtype-specific expression programs in gastric cancer. Cancer Discov 12(3):670–691. https://doi.org/10.1158/2159-8290.CD-21-0683
Rosenbluth JM, Schackmann RCJ, Gray GK, Selfors LM, Li CM-C, Boedicker M et al (2020) Organoid cultures from normal and cancer-prone human breast tissues preserve complex epithelial lineages. Nat Commun 11(1):1711. https://doi.org/10.1038/s41467-020-15548-7
Yang L, Chan AKN, Miyashita K, Delaney CD, Wang X, Li H et al (2021) High-resolution characterization of gene function using single-cell CRISPR tiling screen. Nat Commun 12(1):4063. https://doi.org/10.1038/s41467-021-24324-0
Doke T, Huang S, Qiu C, Liu H, Guan Y, Hu H et al (2021) Transcriptome-wide association analysis identifies DACH1 as a kidney disease risk gene that contributes to fibrosis. J Clin Investig. https://doi.org/10.1172/JCI141801
Chen M, Mao A, Xu M, Weng Q, Mao J, Ji J (2019) CRISPR-Cas9 for cancer therapy: opportunities and challenges. Cancer Lett 447:48–55. https://doi.org/10.1016/j.canlet.2019.01.017
Moncada R, Barkley D, Wagner F, Chiodin M, Devlin JC, Baron M et al (2020) Integrating microarray-based spatial transcriptomics and single-cell RNA-seq reveals tissue architecture in pancreatic ductal adenocarcinomas. Nat Biotechnol 38(3):333–342. https://doi.org/10.1038/s41587-019-0392-8
Sun Y-F, Wu L, Liu S-P, Jiang M-M, Hu B, Zhou K-Q et al (2021) Dissecting spatial heterogeneity and the immune-evasion mechanism of CTCs by single-cell RNA-seq in hepatocellular carcinoma. Nat Commun 12(1):4091. https://doi.org/10.1038/s41467-021-24386-0
Wang S, Rong R, Yang DM, Fujimoto J, Yan S, Cai L et al (2020) Computational staining of pathology images to study the tumor microenvironment in lung cancer. Cancer Res 80(10):2056–2066. https://doi.org/10.1158/0008-5472.CAN-19-1629
Fan J, Slowikowski K, Zhang F (2020) Single-cell transcriptomics in cancer: computational challenges and opportunities. Exp Mol Med 52(9):1452–1465. https://doi.org/10.1038/s12276-020-0422-0
Welch JD, Kozareva V, Ferreira A, Vanderburg C, Martin C, Macosko EZ (2019) Single-cell multi-omic integration compares and contrasts features of brain cell identity. Cell. https://doi.org/10.1016/j.cell.2019.05.006
Stuart T, Satija R (2019) Integrative single-cell analysis. Nat Rev Genet 20(5):257–272. https://doi.org/10.1038/s41576-019-0093-7
Dai H, Li L, Zeng T, Chen L (2019) Cell-specific network constructed by single-cell RNA sequencing data. Nucleic Acids Res 47(11):e62. https://doi.org/10.1093/nar/gkz172
Li H, Courtois ET, Sengupta D, Tan Y, Chen KH, Goh JJL et al (2017) Reference component analysis of single-cell transcriptomes elucidates cellular heterogeneity in human colorectal tumors. Nat Genet 49(5):708–718. https://doi.org/10.1038/ng.3818
Malta TM, Sokolov A, Gentles AJ, Burzykowski T, Poisson L, Weinstein JN et al (2018) Machine learning identifies stemness features associated with oncogenic dedifferentiation. Cell. https://doi.org/10.1016/j.cell.2018.03.034
Venteicher AS, Tirosh I, Hebert C, Yizhak K, Neftel C, Filbin MG et al (2017) Decoupling genetics, lineages, and microenvironment in IDH-mutant gliomas by single-cell RNA-seq. Science. https://doi.org/10.1126/science.aai8478
Zhang Y, Song J, Zhao Z, Yang M, Chen M, Liu C et al (2020) Single-cell transcriptome analysis reveals tumor immune microenvironment heterogenicity and granulocytes enrichment in colorectal cancer liver metastases. Cancer Lett 470:84–94. https://doi.org/10.1016/j.canlet.2019.10.016
Li H, van der Leun AM, Yofe I, Lubling Y, Gelbard-Solodkin D, van Akkooi ACJ et al (2019) Dysfunctional CD8 T cells form a proliferative, dynamically regulated compartment within human melanoma. Cell. https://doi.org/10.1016/j.cell.2018.11.043
Liu J, Adhav R, Miao K, Su SM, Mo L, Chan UI et al (2020) Characterization of BRCA1-deficient premalignant tissues and cancers identifies Plekha5 as a tumor metastasis suppressor. Nat Commun 11(1):4875. https://doi.org/10.1038/s41467-020-18637-9
Satpathy AT, Granja JM, Yost KE, Qi Y, Meschi F, McDermott GP et al (2019) Massively parallel single-cell chromatin landscapes of human immune cell development and intratumoral T cell exhaustion. Nat Biotechnol 37(8):925–936. https://doi.org/10.1038/s41587-019-0206-z
Wu AA, Bever KM, Ho WJ, Fertig EJ, Niu N, Zheng L et al (2020) A phase ii study of allogeneic GM-CSF-transfected pancreatic tumor vaccine (GVAX) with ipilimumab as maintenance treatment for metastatic pancreatic cancer. Clin Cancer Res 26(19):5129–5139. https://doi.org/10.1158/1078-0432.CCR-20-1025
Cadot S, Valle C, Tosolini M, Pont F, Largeaud L, Laurent C et al (2020) Longitudinal CITE-Seq profiling of chronic lymphocytic leukemia during ibrutinib treatment: evolution of leukemic and immune cells at relapse. Biomark Res 8(1):72. https://doi.org/10.1186/s40364-020-00253-w
Zheng B, Wang D, Qiu X, Luo G, Wu T, Yang S et al (2020) Trajectory and functional analysis of PD-1 CD4CD8 T cells in hepatocellular carcinoma by single-cell cytometry and transcriptome sequencing. Adv Sci (Weinh) 7(13):2000224. https://doi.org/10.1002/advs.202000224
Gubin MM, Esaulova E, Ward JP, Malkova ON, Runci D, Wong P et al (2018) High-dimensional analysis delineates myeloid and lymphoid compartment remodeling during successful immune-checkpoint cancer therapy. Cell. https://doi.org/10.1016/j.cell.2018.09.030
Azizi E, Carr AJ, Plitas G, Cornish AE, Konopacki C, Prabhakaran S et al (2018) Single-cell map of diverse immune phenotypes in the breast tumor microenvironment. Cell. https://doi.org/10.1016/j.cell.2018.05.060
Ji AL, Rubin AJ, Thrane K, Jiang S, Reynolds DL, Meyers RM et al (2020) Multimodal analysis of composition and spatial architecture in human squamous cell carcinoma. Cell. https://doi.org/10.1016/j.cell.2020.05.039
Aoki T, Chong LC, Takata K, Milne K, Hav M, Colombo A et al (2020) Single-cell transcriptome analysis reveals disease-defining T-cell subsets in the tumor microenvironment of classic hodgkin lymphoma. Cancer Discov 10(3):406–421. https://doi.org/10.1158/2159-8290.CD-19-0680
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
ZQL, XWH, and QD provided direction and guidance throughout the preparation of this manuscript. HYL, ZQL, and QD wrote and edited the manuscript. QD reviewed and made significant revisions to the manuscript. SYW, MJD, JXL, and QD collected and prepared the related papers. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, Z., Li, H., Dang, Q. et al. Integrative insights and clinical applications of single-cell sequencing in cancer immunotherapy. Cell. Mol. Life Sci. 79, 577 (2022). https://doi.org/10.1007/s00018-022-04608-4
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-022-04608-4