Abstract
Groundwater is an important freshwater resource and hosts specialized microbial assemblages providing fundamental ecosystem services. The current knowledge on the role of aquatic microorganisms in subsurface ecosystems is still limited. This work aimed to explore the links between groundwater hydrogeochemical properties and microbial community traits in a volcanic unconfined aquifer, moderately impacted by anthropic activities. The main physical and chemical parameters of groundwater samples were analyzed, along with microbial biomass (total cell counts, ATP-active biomass concentration), potential metabolic activity, and physiological profiles at the microbial community level (Biolog ™ EcoPlates). The results showed the coexistence of oxidizing and reducing groundwater conditions across the study area. We discriminated two groups of oxidizing/reducing groundwater samples (Ox and Red), each including two subgroups with different chemical conditions attributed to contrasting levels of anthropogenic impact for non-intensive agricultural practices and waste disposal activities (Ox − and Ox + ; Red − and Red +). Although the microbial biomass was likely not affected by changing redox, the microbial metabolic potential and functional diversity changed significantly. In the Ox samples, the community-level physiological profiles were different, mainly owing to the utilization of carboxylic acids (Ox − > Ox +) and carbohydrates (Ox + > Ox −). In the Red samples, a wider set of organic substrates were consumed by the microbial communities, including those less bioavailable (e.g., phenols). Significant differences were also found between Red − and Red + , mainly owing to the relative increase in the utilization of amino acids in Red − , polymers and amines in Red + , along with the active biomass. By reflecting the local redox conditions and moderate levels of anthropogenic impact, the applied approach highlighted changes of microbial metabolic potential and physiological profiles that imply direct repercussions on biogeochemical cycling and the ecosystem services provided by groundwater microorganisms.
Similar content being viewed by others
Introduction
Being protected from surface environmental changes by layers of rocks and unconsolidated materials, aquifers are groundwater-dependent ecosystems that have been persisted relatively unaltered for million years (Gibert and Deharveng 2002). Natural and anthropogenic factors of different origin, extent, and intensity can affect the pristine aquifer conditions, with direct consequences on ecological functioning and ecosystem services provided by the resident biota (Danielopol and Griebler 2008; Battisti et al. 2016; Burri et al. 2019; Ma et al. 2019; Banitz et al. 2020). Current regulation uses fecal indicators as microbial pollution compliance parameters to determine the groundwater quality status intended for human consumption, while a broader environmental perspective should be considered to assess the aquifer ecological status (Griebler and Avramov 2015). This gap represents a crucial issue to gain a holistic scenario, as suggested since more than a decade ago (Gibert et al. 2009).
A better understanding of the groundwater ecology is crucial, since surface waters are becoming increasingly unreliable as a result of higher pollution levels and more frequent climate-related anomalies (e.g., floods and droughts). Aquifers host specialized biological community assemblages, in which microbes provide fundamental ecosystem services by regulating all biogeochemical cycles and maintaining the good groundwater quality status. The groundwater microbial communities promptly respond to environmental changes and the assessment of their taxonomic and metabolic profiles allow relating the community dynamics to the ecosystem status as a whole (Lin et al. 2012; Flynn et al. 2013; Ma et al. 2019). The taxonomy-based approach is the most frequently applied owing to well-established workflows for data elaboration to characterize the phylogenetic composition of the aquatic microbial community. The functional-based approach can facilitate the handling and interpretation of microbial diversity patterns by measuring functional traits across environmental gradients and under the specific conditions a given community is exposed to (Krause et al. 2014; Escalas et al. 2019; Malik et al. 2020). Therefore, the ecosystem responsiveness emerges as a result of the interactions between individuals and the surrounding environment, along with the spatial and temporal distribution of microorganisms with different metabolic potential that likely depends on their functional traits rather than on their taxonomical affiliation (Kiørboe et al. 2018). A critical environmental issue is based on the evidence that changing microbial activities can affect the local redox conditions, thus promoting the mobilization of elements of human health concern in the groundwater resources (e.g., Fe, Mn, As) (Paul et al. 2015).
The ecosystem functioning research is attracting a growing consensus on the need for more functionality-oriented trait-based approaches (Wallenstain and Hall 2012; Litchman et al. 2015). This perspective can offer opportunities for a deeper mechanistic understanding of the role of microbial functional biodiversity in maintaining multiple ecosystem processes and services in all aquatic environments (Krause et al. 2014). Compared to other surface (e.g., lotic and lentic systems) and subsurface freshwater environments (e.g., karstic and alluvial systems), the current knowledge on the microbial ecology of shallow volcanic aquifers is still limited, given the inadequacy of available data and the few dedicated researches (Amalfitano et al. 2014; Segawa et al. 2015; Merino et al. 2022).
The objective of this study was to explore the links between the groundwater hydrogeochemical properties and the functional traits of the aquatic microbial community in a volcanic unconfined aquifer, moderately impacted by non-intensive agricultural practices and waste management plants (i.e., two managed sites for the treatment and disposal of non-hazardous waste). We hypothesize that geogenic local conditions and the anthropogenic impairment of the groundwater quality will reflect on resident microbial community traits, assessed by rapid characterization methods (flow cytometry, ATP-metry, and BiologTMEcoPlates).
Materials and methods
Study area and field sampling
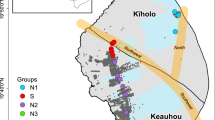
The sampling campaign was performed in a rural area of 60 km2, located in central Italy (municipalities of Viterbo and Vitorchiano, Northern Latium) (Fig. 1a). The target area is adjacent to protected sites of the European Community interest, such as the Lake Bolsena, the largest volcanic lake in Europe. The unconfined aquifer, hosted in the Pleistocene high-K alkaline volcanic products of the Vulsino district (Peccerillo 2005), is highly fractured (Di Filippo et al 1999). The aquifer lies on a low-permeability cover of Flysch units (Upper Cretaceous–Oligocene) and Plio-Pleistocene sediments, under which a geothermal reservoir is hosted in a Mesozoic carbonatic sequence. This setting gives rise to numerous thermal springs in the region (Cinti et al 2014). The water table aquifer is overlain by a permeable unsaturated zone that makes it relatively vulnerable to both diffuse and point source pollution. Depth to water table ranges from few meters to several tens of meters. The anthropogenic impact is mostly due to not intensive farming activities (Fig. 1b) and limited industrial activities, including two sites for the treatment and disposal of non-hazardous waste (Le Fornaci and Casale Bussi plants) (Fig. 1a). Groundwater flow direction is mainly oriented towards S in the Le Fornaci area, with an eastward rotation in the Casale Bussi sector (Capelli et al. 2012; Parrone et al. 2021).
a Sampling sites and piezometric levels (Filled Red dots = Casale Bussi landfill; empty red dots = Le Fornaci landfill); b Land use of the study area, CORINE Land cover 2012 (from http://wms.pcn.minambiente.it/ogc?map=/ms_ogc/wfs/Corine_Land_Cover2012.map—18/02/2022); c Piper diagram of the groundwater samples (n = 28) (Ox − = orange; Ox + = red; Red − = pale blue; Red + = Blue) (color figure online)
The sampling campaign was conducted during the dry season (June–July 2017), characterized by a general reduction in precipitations (Pierangioli et al. 2017). The depth of the selected wells varied between 28.5 and 160.0 m below the ground level and the depth to water table from 7 to 90 m with a median value of 25 m (Table S1). Groundwater samples (n = 28) were withdrawn from private wells utilized for irrigation, three piezometers located inside the Casale Bussi (CB) waste management plant and two piezometers inside Le Fornaci (LF) landfill (Fig. 1a), one of which (LF000) is located up-gradient the landfill.
All samples were collected and transported following standard protocols (Frollini et al. 2019; Preziosi et al. 2019). Briefly, a flow cell equipped with AQUAREAD probes for dissolved oxygen (DO), oxidation–reduction potential (ORP), temperature (T), pH and electrical conductivity (EC), was set for in-line measurements directly from the sampling pipe. After parameters stabilization (usually 30 min), samples were collected through an in-line device to avoid interaction with atmospheric oxygen. All samples were stored in HDPE bottles previously washed with HNO3 and thoroughly rinsed with Milli-Q water.
Samples for anions, cations, and trace elements were filtered onto 0.45-µm pore size polycarbonate filters. After filtration, samples for cation and trace elements analysis were acidified on site (HNO3). Samples for anions were analyzed within 24 h, whereas samples for cations and trace elements within one month. Samples for dissolved organic carbon (DOC) analysis were filtered (0.7 µm pore size glass microfiber filters) and acidified (HCl) on site.
Samples for the analyses of fecal contamination were collected in 100 mL sterile polystyrene bottles provided by the producer and analyzed in the laboratory within 6 h. Samples for the analysis of the prokaryotic cell abundance were collected in 2 mL Eppendorf safe-lock tubes, fixed with formaldehyde (2%, final concentration) and analyzed within 24 h. Samples for the ATP analysis were collected in 5 L glass bottles and analyzed within 24 h. Finally, samples for the analysis of the microbial functional properties were collected in sterile 50 mL Falcon tubes and analyzed within 24 h. All the glassware was accurately washed with HCl (1 N) and rinsed with Milli-Q water.
Groundwater physical–chemical characteristics
The concentration of NO3−, PO43−, SO42−, F− and Cl− was determined by Ionic Chromatography (Dionex DX-120) and the HCO3− concentration was analyzed by HCl potentiometric titration. Major cations and trace elements (Table S1) were analyzed by ICP-Mass Spectrometry (Agilent technologies 7500c). The analytical performance of ICP-MS was tested against a certified material (NIST 1640a, trace elements in natural waters). Dissolved organic carbon (DOC) concentration was measured by TOC analyzer (Shimadzu TOC-5000A analyzer).
Microbial community characterization
The occurrence of fecal contamination was examined by the measurement of the indicator bacteria Escherichia coli (Colilert-18 test kit, DTS, IDEXX Laboratories Inc.) (APHA; AWWA; WEF 2009). The UV-light-positive wells for the activities on 4-methyl-umbelliferil-B-D-glucuronide were counted and referred to the Quanti-Tray/2000 Most Probable Number (MPN) table.
The prokaryotic cell abundance was analyzed by the flow cytometer A50-micro (Apogee Flow System, Hertfordshire, UK) equipped with a solid-state laser set at 20 mV and tuned to an excitation wavelength of 488 nm. All samples were stained with SYBR Green I (1:10,000 dilution; Molecular Probes, Invitrogen) for 10 min in the dark at room temperature. The light scattering signals (forward and side scatters), the green fluorescence (530/30 nm) and red fluorescence (> 610 nm) were acquired with a threshold sets on the green channel. Samples were run at low flow rates to keep the number of events below 1000 event s−1. The data were analyzed using the Apogee Histogram Software (v89.0). The total cell counts were determined by the signatures in a plot of the side scatter vs the green fluorescence. The intensity of green fluorescence emitted by SYBR positive cells allowed discriminating two cell groups with low (LNA) and high (HNA) nucleic acid content (Amalfitano et al. 2014). Total cell count values were converted to carbon equivalents by using the factor 25 fg of C per cell (Griebler et al. 2002).
The active microbial biomass was estimated by measuring adenosine triphosphate (ATP) concentration by filtering 2–5 L through 0.2 µm polycarbonate membranes (diameter 45 mm). The whole filter was extracted at room temperature with dimethyl sulfoxide and successively MOPS buffer (10 mM, EDTA 2 mM, pH 7.40) was added (Jakubczak 1980; Puddu et al. 1997). The extract was stored at -20 °C and thawed immediately prior to analysis. The ATP extract was assayed using the firefly luciferase–luciferin enzyme (FLE-250 SIGMA) bioluminescence assay utilizing the luminometer Victor X3 multiplate reader 2030 (Perkin Elmer). The limit of ATP detection was 0.04 nmol mL−1. ATP concentrations were converted to carbon equivalent using the factor 250 mg of C per mg of ATP (Karl 1980).
Biolog EcoPlates assay and the Community-Level Physiological Profile (CLPP)
The Biolog™ EcoPlates assay (Biolog, Inc., Hayward, California, USA) consists of 96 micro-wells containing 31 organic carbon substrates labeled with the respiration-sensitive tetrazolium dye plus one control (each plate contains all substrates and control in three replicates). All the groundwater samples underwent the same treatment. Within 6 h from sampling, 150 µL of groundwater sample was introduced in each micro-well of the EcoPlates in triplicate. The plates were covered and incubated at 20 °C in the dark. The samples belonging to oxidizing and reducing faces were incubated at the same conditions considering that the DO concentration was always above 0.1 mg L−1 with the only exception of two samples (Table S1). This approach made possible the comparisons of the results. After 24 h of incubation, the color developed was detected at the wavelength 590 nm by a multi-label plate reader (PerkinElmer VICTOR™ X3). An incubation time of 24 h was selected to explore the prompt response of the microbial functional traits to the available organic substrates. The values of optical density (OD) at 24 h were subtracted by those found at 0 h to eliminate the blank absorbance signal (Insam et al. 2004). Negative values were set to zero. The microbial metabolic potential was calculated as the mean degradation activity on all 31 substrates provided and defined as Average Well Color Development (AWCD), (Garland et al. 1991). For each sample, OD590 nm data were previously normalized by relative AWCD values. The substrates provided were grouped into six chemical categories (i.e., carbohydrates, polymers, carboxylic acids, phenolic compounds, amino acids, and amines). The contribution of each class to the total degradative activity was calculated. The community-level physiological profile (CLPP) was estimated by the analysis of each single substrate utilized by the microbial community. The functional diversity was assessed through the application of the Shannon–Wiener’s diversity index (H) (Melita et al. 2019).
Statistical analyses
Since most variables did not exhibit a normal data distribution, both univariate and multivariate statistics were performed using non-parametric tests. In order to compare different types of variables (i.e., physical, chemical and microbial parameters), a standardization was performed on the entire data set by applying the log-transformation \(\mathrm{log }\left(1+x\right)\).
The multivariate ANalysis Of SIMilarities (ANOSIM test), based on the Gower (dis)similarity matrix, was used to test statistical differences among the sample groups and subgroups by considering all physical, chemical, and microbial parameters (Tables S1 and S2). The univariate non-parametric Kruskal–Wallis test for equal medians, along with the Mann–Whitney pairwise comparison, was performed to assess differences between sample groups per each single parameter. A multi-group SIMilarity PERcentage test (SIMPER), using the Gower (dis)similarity measure (multiplied by 100), was run to identify the parameters that were primarily responsible for observed differences between the sample groups.
The graphical representation of (dis)similarities among groups was obtained through the nMDS ordination plot, based on the Gower (dis)similarity matrix. Statistical comparisons on degradative activity of single substrates were performed through a Non-Parametric Multivariate ANalysis Of VAriance (NPMANOVA), based on the Gower (dis)similarity matrix.
Statistical analyses and graphical representations were performed using the software PAST v4.0 (Hammer et al. 2001).
Results
Physical and chemical characteristics of the groundwater samples
The hydrogeochemical characterization identified a calcium-bicarbonate water type with few sulfate–chloride type samples (Fig. 1c). The NMDS analysis (Fig. 2), performed on 32 physical and chemical parameters measured on 28 groundwater samples (Table S1), showed two statistically different groups of samples (ANOSIM test, p < 0.01), collected at oxidizing (Ox, n = 19) and reducing redox conditions (Red, n = 9). Between the two sample groups, we found significant differences for 22 out of 32 tested parameters (Kruskal–Wallis test, p < 0.05), with total dissimilarity explained mainly by Mn, Fe and V, DO, NO3−, and SO42−concentrations (SIMPER test). Moreover, two subgroups were identified according to the chemical characteristics (Ox − vs Ox + , Red − vs Red +). Ox + (n = 7) showed relatively higher concentrations of NO3−, PO43−, SO42− and B than Ox − (n = 12). Red + (n = 5) showed relatively higher concentrations of Cl−, K, Ni, Cu and DOC than Red − (n = 4) (Table 1). By considering the threshold values imposed by current regulations (NO3− = 50 mg L−1; Cl− = 250 mg L−1), the concentration of nitrates in Ox + (mean value 57.3 ± 24.4 mg L−1) and chloride in Red + (mean value 152.0 ± 111.0 mg L−1) suggested a moderate anthropogenic impairment of the groundwater quality (Table S1), as expected in a rural area and in proximity of landfill plants. Only in the site LF000, belonging to the Red + group and sited upstream the landfill, Cl− concentration exceeded the given threshold (Table S1).
Microbial biomass and functional characteristics
The occurrence of E. coli was found in 7 out of 28 total samples, without a clear pattern among sample groups (range 1–54.8 MPN 100 mL−1) (Table S2). Similarly, the total cell counts and HNA/LNA cell ratio did not change significantly among groups (Table S2).
The total prokaryotic biomass (C-PB) ranged from 0.13 to 7.49 nmol C mL−1 (corresponding to an average value of 1.1 × 105 cells mL−1), with higher values found in the Red + subgroup (Fig. 3a). The active biomass concentration (C-ATP) ranged from 0.01 to 1.32 nmol C mL−1 (Fig. 3b), with significantly higher values in the Red + than the other subgroups (median C-ATP 0.33 nmol L−1; p < 0.05).
Microbial community properties in samples collected from oxidizing (Ox) and reducing (Red) redox groundwater conditions at contrasting impact levels (Ox − vs Ox + , Red − vs Red +). C-PB Prokaryotic biomass; C-ATP ATP-Active Biomass; AWCD Average Well Color Development; S Shannon–Wiener’s diversity index. Statistical comparison was performed among groups and superscript letters indicate statistical differences (Mann–Whitney pairwise test; p < 0.05)
The AWCD values varied consistently among samples, from 0 to 0.267 OD590nm (i.e., site CB3 of Red +), with higher values in Red − and Red + than Ox − and Ox + (Fig. 3c, Table S2). The functional diversity, based on the utilization of the substrates provided by the Biolog test, showed significant lower values in Ox than Red sample groups (i.e., Ox − < Red − ; Ox + < Red + ; p < 0.05) (Fig. 3d).
Community-level physiological profiles
The microbial community in Ox − showed the lowest number of reactive wells. Seven out 31 organic substrates provided by the Biolog test were not utilized in the Ox − group, whereas only three were not utilized in the Ox + group (Table 2). In Red, the number of reactive wells increased significantly with respect to Ox and all substrates were utilized at a relatively higher degradation rate (Table 2). The multivariate analysis highlighted differences in the CLPP patterns between Ox and Red groups (Fig. 4).
In Ox − , the utilization of carboxylic acids was significantly higher (p < 0.05) and contributed on average to the 56% of the total potential metabolic activity (Fig. 5). In particular, the γ-hydroxybutyric acid resulted the most degraded carboxylic acid, explaining 27% of the total dissimilarity between Ox − and Ox + (Simper test). A decreasing contribution to the total metabolic activity was observed for amino acids (19%), carbohydrates (17%), polymers (7%), and amines (1%), with no reaction found on both phenolic compounds (Fig. 5).
Contribution of classes of substrates to the total microbial metabolic potential (OD590nm) in the oxidizing (Ox – and Ox +) and reducing (Red – and Red +) redox conditions. Statistical comparison was performed among groups and superscript letters indicate statistical differences (Mann–Whitney pairwise test; p < 0.05)
Ox + metabolic activity was mainly own to carbohydrates (38%), carboxylic acids (19%), amines (18%), polymers (13%), amino acids (12%), and phenolic compounds (< 1%) (Fig. 5). In particular, i-erythritol, putrescine, and γ-hydroxybutyric acid contributed on average for more than the 10% to the total absorbance (Fig. 4, Table 2). Within carbohydrates, i-erythritol and N-acetyl-D-glucosamine were on average, preferentially utilized in place of cellobiose, and a better utilization of the putrescine was observed (from 1% in Ox − to 18% in Ox +).
In Red, the substrate utilization patterns of the two subgroups were similar, with carbohydrates (i.e., i-erythritol) and carboxylic acids (i.e., γ-hydroxybutyric acid and itaconic acid) as the most utilized substrates (Fig. 5). The microbial metabolism was also fueled by phenolic compounds, showing significantly higher absorbance values with respect to Ox (p < 0.01). In Red − , the average contribution of amino acids to the total absorbance increased to 24%, mainly supported by L-asparagine and L-threonine (Fig. 5, Table 2). In Red + , a relatively higher contribution was found for polymers (13%) and amines (7%) (Fig. 5).
Discussion
The groundwater quality is strictly dependent on hydrological conditions, lithological settings, and inputs from the surface (Descourvieres et al. 2010; Preziosi et al. 2019). The investigated aquifer is located in a volcanic area where significant vertical and lateral lithological variation leads to variable hydrodynamic properties. Different geogenic processes (e.g., water–rock interaction, upwelling of geothermal fluids, and presence of hydrothermal mineralization) can contribute to the local enrichment of specific elements (e.g., As and F) and high spatial variability of hydrogeochemical characteristics of groundwater (Parrone et al. 2020). The study area is characterized by the co-presence of oxidizing and reducing facies with different areal distribution (Parrone et al. 2021). In reducing conditions, the reductive dissolution of oxi-hydroxides likely induced high Fe and Mn concentrations (Preziosi et al. 2019; Parrone et al. 2021). The relatively high nutrient concentrations (i.e., NO3− and PO43−) observed in Ox + can originate from the agricultural practices (Narany et al. 2017; Serio et al. 2018; Liu et al. 2021). The relatively high Cl− concentrations that characterized Red + could indicate either a landfill-induced pollution (Aharoni et al. 2020) or geogenic sources, given the abundant brines and Triassic evaporites found in the Vulsini region (Frondini et al. 2012). Despite further evidences are needed, the impact of the landfill sites appeared only moderate and the high Cl− concentrations also found upstream (i.e., sample LF000) could suggest a concomitant geogenic source.
The variable pattern of fecal contamination among the sample groups suggested external inputs from surface point sources regardless the local hydrogeochemical conditions, as expected in an unconfined aquifer. However, the active metabolism of the groundwater microbial community, primarily responsible for catalyzing diverse oxidation/reduction reactions and controlling the release of minerals, fate and transport of metals and organic compounds (Kirk et al. 2004; Song et al. 2007), might have contributed to shape the chemical profiles of the target aquifer. The microbially driven release of microelements may occur with the potential to rendering some metals readily available for uptake and accumulation in plants and animals (i.e., Mn and Fe) (Kraemer and Hering 2004; Chapelle et al. 2012; Adekunle et al. 2020). Moreover, carbon oxidation is described to play a key control on the speciation and transport of redox-sensitive elements (e.g., Mn/Fe/S redox speciation) and the proliferation of specific microbial taxa (Percak-Dennet and Roden 2014; Stegen et al. 2016).
In this study, different redox conditions and a moderate anthropogenic impairment of the groundwater quality directly reflected on microbial functional traits. The microbial community resident in the oxidizing redox facies showed a metabolic profile reported for resource limited and low energy environment, characterized by a relatively low prokaryotic biomass, metabolic potential and functional diversity with respect to that in the reducing facies (Griebler and Lueders 2009). In Ox − , the low affinity to most of the Biolog substrates could indicate a relatively low metabolic versatility of the microbial community to adapt to newly available substrates, as previously reported for subsurface aquatic environments in pristine conditions (Griebler and Avramov 2015; Melita et al. 2019). This result might imply a relatively high vulnerability of this ecosystem, since the resident microbial communities are likely unable to degrade complex organic compounds they may accidentally enter in contact with (e.g., organic pollutants). On the contrary, reducing redox conditions were likely to promote relatively higher metabolic potential and functional diversity, along with a higher affinity for all organic substrates and metabolic pathways. Notably, the microbial community could exploit a wider suite of carbon sources using all available electron acceptors (e.g., ferric iron, manganese, sulfate) under suboxic conditions (Kirk et al. 2004; Song et al. 2007; Janniche et al. 2012). The groundwater quality impairment can induce significant CLPP modifications (Melita et al. 2019; Pradeep Ram et al. 2020). In Ox − , the microbial community showed a better ability to feed on energy-rich carbon substrates (i.e., carboxylic acids) with respect to Ox + , although no changes were observed in the prokaryotic biomass and the potential metabolic activity.
Significant differences were observed in prokaryotic traits between Red − and Red + . In Red + , microbial community showed a relatively higher active biomass concentration (C-ATP) along with the ability to utilize complex organic substrates as energy sources, including polymers and amines. Similar findings were reported previously for impacted areas (Broholm and Arvin 2000; Sanchez-Maldonado et al. 2011; Hedbavna et al. 2016; Fillinger et al. 2019). The ability in consuming structurally complex substrata, might indicate altered conditions where the microbial community is naturally adapted to feed on these substances (Oest et al. 2018; Melita et al. 2019). Indeed, the microbial degradation of complex polymers (i.e., glycogen, tween 40, tween 80) implies the presence of specific microbial functional traits and extracellular enzymes (Henrissat et al. 2002; Tamaki et al. 2003). The significant consumption of N-rich substrata (amines) observed in Red + is compatible with N-depletion of the microbial degradative processes also considering the relatively high activity occurring with respect to Red − .
The Biolog EcoPlates assay is a relatively simple and inexpensive technique for comparing microbial communities by rapidly providing reliable responses on their functional properties. However, several methodological constraints for environmental applications are reported in the specialized literature (Preston-Mafham et al. 2002; Christian and Lind 2006). In particular, only few of the 31 organic substrates provided are present in the natural environments; thus, the microbial degradative responses will not necessarily reflect the on-site community functions (Garland 1999). However, it was noted that the use of less common (rather than ecologically relevant) organic substrates provided more discriminatory power to rapidly detect microbial community changes through the CLPP analysis (Konopka et al. 1998).
Our findings highlight consistent differences in the microbial functional traits that may play a role in attenuating the effects of anthropogenic alterations (Bhatt et al. 2020), but also imply direct consequences on biogeochemical cycles (e.g., fast carbon and nutrient cycling, biodegradation of anthropogenic contaminants) and groundwater quality (e.g., oxygen depletion, speciation, and transport of metals).
Conclusions
Ecologically oriented studies can provide insights into the biochemical dynamics driven by groundwater biota and open new perspectives into the subsurface environmental conservations. Evaluating the effects of natural and anthropogenic factors on the biota represents a challenge for groundwater ecologists, mostly to discriminate, quantify, and counteract the potential consequences for the environment. This field study provided a snapshot of the microbial community physiological profiles and highlighted significant and rapid microbial metabolic adaptations to conditions imposed by natural (i.e., redox conditions) and anthropogenic factors of moderate impact level (i.e., nutrient enrichment). The adopted functional trait-based approach was proved to be a suitable tool to promptly assess the effects of environmental modifications on microbial metabolic activities directly involved in nutrient cycling, pollutant degradation, and the overall ecosystem services provided by groundwater microorganisms.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Adekunle AS, Oyekunle JAO, Ojo OS et al (2020) Heavy metal speciation, microbial study and physicochemical properties of some groundwaters: a case study. Chem Africa 3:211–226. https://doi.org/10.1007/s42250-019-00099-2
Aharoni I, Siebner H, Yogev U, Dahan O (2020) Science of the total environment holistic approach for evaluation of land fi ll leachate pollution potential – from the waste to the aquifer. Sci Total Environ 741:140367. https://doi.org/10.1016/j.scitotenv.2020.140367
APHA, AWWA, & WEF. (2009) Standard methods for the examination of water and wastewater. In: Eaton AD, Clesceri LS, Greenberg AE (eds) 21st edn (on-line). APHA, Washington, DC
Banitz T, Chatzinotas A, Worrich A (2020) Prospects for integrating disturbances, biodiversity and ecosystem functioning using microbial systems. Front Ecol Evol 8:1–9. https://doi.org/10.3389/fevo.2020.00021
Battisti C, Poeta G, Fanelli G (2016) Role and effects of disturbances in natural systems BT - an introduction to disturbance ecology: a road map for wildlife management and conservation. In: Poeta G, Fanelli G (eds) Battisti C. Springer International Publishing, Cham, pp 13–29
Bhatt P, Gangola S, Bhandari G et al (2020) New insights into the degradation of synthetic pollutants in contaminated environments. Chemosphere 268:128827
Broholm MM, Arvin E (2000) Biodegradation of phenols in a sandstone aquifer under aerobic conditions and mixed nitrate and iron reducing conditions. J Contam Hydrol 44:239–273
Burri NM, Weatherl R, Moeck C, Schirmer M (2019) A review of threats to groundwater quality in the anthropocene. Sci Total Environ 684:136–154. https://doi.org/10.1016/j.scitotenv.2019.05.236
Capelli G, et al (2012). Carta idrogeologica del territorio della Regione Lazio. 1:100.000 (4 fogli), Regione Lazio, S.EL.CA. Firenze
Chapelle FH, Bradley PM, Mcmahon PB et al (2012) Dissolved oxygen as an indicator of bioavailable dissolved organic carbon in groundwater. Ground Water 50:230–241. https://doi.org/10.1111/j.1745-6584.2011.00835.x
Christian BW, Lind OT (2006) Key issues concerning Biolog use for aerobic and anaerobic freshwater bacterial community-level physiological profiling. Int Rev Hydrobiol 91(3):257–268
Cinti D, Tassi F, Procesi M et al (2014) Fluid geochemistry and geothermometry in the unexploited geothermal field of the Vicano-Cimino Volcanic District (Central Italy). Chem Geol 371:96–114. https://doi.org/10.1016/j.chemgeo.2014.02.005
Danielopol DL, Griebler C (2008) Changing paradigms in groundwater ecology – from the ‘living fossils’ tradition to the ‘new groundwater ecology.’ Int Rev Hydrobiol 93:565–577. https://doi.org/10.1002/iroh.200711045
Descourvieres C, Prommer H, Oldham C et al (2010) Kinetic reaction modeling framework for identifying and quantifying reductant reactivity in heterogeneous aquifer sediments. Environ Sci Technol 44:6698–6705. https://doi.org/10.1021/es101661u
Di Filippo M, Lombardi S, Nappi G et al (1999) Volcano-tectonic structures, gravity and helium in geothermal areas of Tuscany and Latium (Vulsini volcanic district), Italy. Geothermics 28:377–393. https://doi.org/10.1016/S0375-6505(99)00014-0
Escalas A, Hale L, Voordeckers JW et al (2019) Microbial functional diversity: from concepts to applications. Ecol Evol 9:12000–12016. https://doi.org/10.1002/ece3.5670
Fillinger L, Hug K, Trimbach AM et al (2019) The D-A-(C) index: a practical approach towards the microbiological-ecological monitoring of groundwater ecosystems. Water Res 163:114902. https://doi.org/10.1016/j.watres.2019.114902
Flynn TM, Sanford RA, Ryu H et al (2013) Functional microbial diversity explains groundwater chemistry in a pristine aquifer. BMC Microbiol 13:1–15
Frollini E, Rossi D, Rainaldi M et al (2019) A proposal for groundwater sampling guidelines: application to a case study in Southern Latium. Rend Online Soc Geol Ital 47:46–51
Frondini F, Cardellini C, Caliro S et al (2012) Regional groundwater flow and interactions with deep fluids in western Apennine: the case of Narni-Amelia chain (Central Italy). Geofluids 12:182–196. https://doi.org/10.1111/j.1468-8123.2011.00356.x
Garland JL, Mills AL (1991) Classification and Characterization of heterotrophic microbial communities on the basis of patterns of community-level sole-carbon-source utilization. Appl Environ Microbiol 57:2351 (LP–2359)
Garland JL (1999) Potential and limitations of BIOLOG for microbial community analysis. In: Proceedings of the 8th international symposium on microbial ecology Atlantic Canada society for microbial ecology. Society for Microbial Ecology, pp 1–7
Gibert J, Deharveng L (2002) Subterranean ecosystems: a truncated functional biodiversity: this article emphasizes the truncated nature of subterranean biodiversity at both the bottom (no primary producers) and the top (very few strict predators) of food webs and discusses the implications of this truncation both from functional and evolutionary perspectives. Bioscience 52:473–481. https://doi.org/10.1641/0006-3568(2002)052[0473:SEATFB]2.0.CO;2
Gibert J, Culver DC, Dole-Olivier MJ et al (2009) Assessing and conserving groundwater biodiversity: synthesis and perspectives. Freshw Biol 54:930–941. https://doi.org/10.1111/j.1365-2427.2009.02201.x
Griebler C, Mindl B, Slezak D, Geiger-Kaiser M (2002) Distribution patterns of attached and suspended bacteria in pristine and contaminated shallow aquifers studied with an in situ sediment exposure microcosm. Aquat Microb Ecol 28:117–129. https://doi.org/10.3354/ame028117
Griebler C, Lueders T (2009) Microbial biodiversity in groundwater ecosystems. Freshw Biol 54:649–677. https://doi.org/10.1111/j.1365-2427.2008.02013.x
Griebler C, Avramov M (2015) Groundwater ecosystem services: a review. Freshw Sci 34:355–367. https://doi.org/10.1086/679903
Hammer Ø, Harper DAT, Ryan PD (2001) PAST: Paleontological statistics software package for education and data analysis. Palaeontol Electron 4:9
Hedbavna P, Rolfe SA, Huang WE, Thornton SF (2016) Biodegradation of phenolic compounds and their metabolites in contaminated groundwater using microbial fuel cells. Bioresour Technol 200:426–434. https://doi.org/10.1016/j.biortech.2015.09.092
Henrissat B, Deleury E, Coutinho PM (2002) Glycogen metabolism loss: a common marker of parasitic behaviour in bacteria? Trends Genet 18:437–440. https://doi.org/10.1016/S0168-9525(02)02734-8
Insam H, Goberna M (2004) Molecular microbial ecology manual. Springer Netherlands, Dordrecht
Jakubczak E (1980) Mesure de l’ATP bactérien par bioluminescence: étude critique des méthodes d’extraction. Ann Biol Clin (paris) 38(5):297–304
Janniche GS, Spliid H, Albrechtsen HJ (2012) Microbial community-level physiological profiles (CLPP) and herbicide mineralization potential in groundwater affected by agricultural land use. J Contam Hydrol 140–141:45–55. https://doi.org/10.1016/j.jconhyd.2012.08.008
Karl DM (1980) Cellular nucleotide measurements and applications in microbial ecology. Microbiol Rev 44:739–796
Kiørboe T, Visser A, Andersen KH (2018) A trait-based approach to ocean ecology. ICES J Mar Sci 75:1849–1863
Kirk JL, Beaudette LA, Hart M et al (2004) Methods of studying soil microbial diversity. J Microbiol Methods 58:169–188
Konopka AL, Oliver L, Turco RF (1998) The use of carbon substrate utilization patterns in environmental and ecological microbiology. Microb Ecol 35:103–115
Kraemer SM, Hering JG (2004) Biogeochemical controls on the mobility and bioavailability of metals in soils and groundwater-preface. Aquat Sci 66:1–2
Krause S, Le Roux X, Niklaus PA et al (2014) Trait-based approaches for understanding microbial biodiversity and ecosystem functioning. Front Microbiol 5:1–10. https://doi.org/10.3389/fmicb.2014.00251
Lin X, McKinley J, Resch CT et al (2012) Spatial and temporal dynamics of the microbial community in the Hanford unconfined aquifer. ISME J 6:1665–1676
Litchman E, Edwards KF, Klausmeier CA (2015) Microbial resource utilization traits and trade-offs: implications for community structure, functioning, and biogeochemical impacts at present and in the future. Front Microbiol 6:254
Liu X, Wang X, Zhang L et al (2021) Impact of land use on shallow groundwater quality characteristics associated with human health risks in a typical agricultural area in Central China. Environ Sci Pollut Res 28:1712–1724
Ma J, Liu H, Tong L et al (2019) Relationships between microbial communities and groundwater chemistry in two pristine confined groundwater aquifers in central China. Hydrol Process 33:1993–2005
Malik AA, Martiny JBH, Brodie EL et al (2020) Defining trait-based microbial strategies with consequences for soil carbon cycling under climate change. ISME J 14:1–9
Melita M, Amalfitano S, Preziosi E et al (2019) Physiological profiling and functional diversity of groundwater microbial communities in a municipal solid waste landfill area. Water (switzerland). https://doi.org/10.3390/w11122624
Merino N, Jackson TR, Campbell JH, Kersting AB, Sackett J, Fisher JC, Moser DP (2022) Subsurface microbial communities as a tool for characterizing regional-scale groundwater flow. Sci Total Environ 842:156768. https://doi.org/10.1016/j.scitotenv.2022.156768
Oest A, Alsaffar A, Fenner M et al (2018) Patterns of change in metabolic capabilities of sediment microbial communities in river and lake ecosystems. Int J Microbiol 2018:1–15. https://doi.org/10.1155/2018/6234931
Parrone D, Ghergo S, Frollini E et al (2020) Arsenic-fluoride co-contamination in groundwater: background and anomalies in a volcanic-sedimentary aquifer in central Italy. J Geochemical Explor 217:106590. https://doi.org/10.1016/j.gexplo.2020.106590
Parrone D, Frollini E, Preziosi E, Ghergo S (2021) eNaBLe, an on-line tool to evaluate natural background levels in groundwater bodies. Water 13:74
Paul D, Kazy SK, Gupta AK, Pal T, Sar P (2015) Diversity, metabolic properties and arsenic mobilization potential of indigenous bacteria in arsenic contaminated groundwater of West Bengal. India Plos One 10(3):e0118735
Peccerillo A (2005) Plio-quaternary volcanism in Italy, vol 365. Springer-Verlag Berlin Heidelberg, New York
Percak-Dennett EM, Roden EE (2014) Geochemical and microbiological responses to oxidant introduction into reduced subsurface sediment from the Hanford 300 Area, Washington. Environ Sci Technol 48:9197–9204. https://doi.org/10.1021/es5009856
Pierangioli L, Cellai G, Ferrise R et al (2017) Effectiveness of passive measures against climate change: case studies in Central Italy. Build Simul 10:459–479. https://doi.org/10.1007/s12273-016-0346-8
Pradeep Ram AS, Keshri J, Sime-Ngando T (2020) Differential impact of top-down and bottom-up forces in structuring freshwater bacterial communities. FEMS Microbiol Ecol. https://doi.org/10.1093/femsec/fiaa005
Preston-Mafham J, Boddy L, Randerson PF (2002) Analysis of microbial community functional diversity using sole-carbon-source utilisation profiles - a critique. FEMS Microbiol Ecol 42(1):1–14. https://doi.org/10.1111/j.1574-6941.2002.tb00990.x (PMID: 19709261)
Preziosi E, Frollini E, Zoppini A et al (2019) Disentangling natural and anthropogenic impacts on groundwater by hydrogeochemical, isotopic and microbiological data: hints from a municipal solid waste landfill. Waste Manag 84:245–255. https://doi.org/10.1016/j.wasman.2018.12.005
Puddu A, La Ferla R, Allegra A et al (1997) Seasonal and spatial distribution of bacterial production and biomass along a salinity gradient (Northern Adriatic Sea). Hydrobiologia 363:271–282
Sánchez-Maldonado AF, Schieber A, Gänzle MG (2011) Structure–function relationships of the antibacterial activity of phenolic acids and their metabolism by lactic acid bacteria. J Appl Microbiol 111:1176–1184
Segawa T, Sugiyama A, Kinoshita T et al (2015) Microbes in groundwater of a volcanic mountain, Mt. Fuji; 16S rDNA phylogenetic analysis as a possible indicator for the transport routes of groundwater. Geomicrobiol J 32(8):677–688
SheikhyNarany T, Aris AZ, Sefie A, Keesstra S (2017) Detecting and predicting the impact of land use changes on groundwater quality, a case study in Northern Kelantan, Malaysia. Sci Total Environ 599–600:844–853. https://doi.org/10.1016/j.scitotenv.2017.04.171
Song J, Fallgren PH, Azra Bilgin A et al (2007) Bioremediation of benzene, ethylbenzene, and xylenes in groundwater under iron-amended, sulfate-reducing conditions. Environ Toxicol Chem 26:249–253
Stegen JC, Konopka A, McKinley JP et al (2016) Coupling among microbial communities, biogeochemistry and mineralogy across biogeochemical facies. Sci Rep 6:30553. https://doi.org/10.1038/srep30553
Tamaki H, Hanada S, Kamagata Y et al (2003) Flavobacterium limicola sp. nov., a psychrophilic, organic-polymer-degrading bacterium isolated from freshwater sediments. Int J Syst Evol Microbiol 53:519–526. https://doi.org/10.1099/ijs.0.02369-0
Wallenstein MD, Hall EK (2012) A trait-based framework for predicting when and where microbial adaptation to climate change will affect ecosystem functioning. Biogeochemistry 109:35–47
Acknowledgements
The present work was financially supported by the Regione Lazio (Area Ciclo Integrato Rifiuti), Italy (Contract n. 6949, prot. n. 5407, 3/12/2018) and by Ecologia Viterbo s.r.l., Italy (contract n.14297, prot. n. 3269, 11-08-2017). Marco Melita was supported by the flagship Project RITMARE (SP3-WP2-AZ1-UO06) coordinated by the National Research Council of Italy and funded by the Italian Ministry of Education, University and Research within the National Research Program 2012-2016. The authors also wish to thank Dr. Laura Amicucci of Geovit s.a.s. for the logistical support during the sampling campaign.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Melita, M., Amalfitano, S., Preziosi, E. et al. Redox conditions and a moderate anthropogenic impairment of groundwater quality reflected on the microbial functional traits in a volcanic aquifer. Aquat Sci 85, 3 (2023). https://doi.org/10.1007/s00027-022-00899-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00027-022-00899-8