Abstract
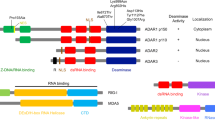
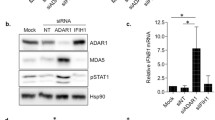
The innate immune system is the first line of the cellular defence against invading pathogens. A critical component of this defence is the capacity to discriminate foreign RNA molecules, which are distinct from most cellular RNAs in structure and/or modifications. However, a series of rare autoimmune/autoinflammatory diseases in humans highlight the propensity for the innate immune sensing system to be activated by endogenous cellular double-stranded RNAs (dsRNAs), underscoring the fine line between distinguishing self from non-self. The RNA editing enzyme ADAR1 has recently emerged as a key regulator that prevents innate immune pathway activation, principally the cytosolic dsRNA sensor MDA5, from inducing interferon in response to double-stranded RNA structures within endogenous RNAs. Adenosine-to-Inosine RNA editing by ADAR1 is proposed to destabilise duplexes formed from inverted repetitive elements within RNAs, which appear to prevent MDA5 from sensing these RNA as virus-like in the cytoplasm. Aberrant activation of these pathways has catastrophic effects at both a cellular and organismal level, contributing to one of the causes of the conditions collectively known as the type I interferonopathies.


Similar content being viewed by others
References
Crow YJ, Manel N (2015) Aicardi-Goutières syndrome and the type I interferonopathies. Nat Rev Immunol 15:429–440
Crow YJ, Chase DS, Schmidt JL, Marcin S, Forte GMA, Gornall HL, Anthony O, Beverley A, Amy P, Guy H et al (2015) Characterization of human disease phenotypes associated with mutations in TREX1, RNASEH2A, RNASEH2B, RNASEH2C, SAMHD1, ADAR, and IFIH1. Am J Med Genet A 167:296–312
Barbalat R, Ewald SE, Mouchess ML, Barton GM (2011) Nucleic acid recognition by the innate immune system. Annu Rev Immunol 29:185–214
Wu J, Jiaxi W, Chen ZJ (2014) Innate immune sensing and signaling of cytosolic nucleic acids. Annu Rev Immunol 32:461–488
Hur S, Sun H (2015) Molecular mechanisms of viral RNA detection: RIG-I and MDA5. Biophys J 108:335a
Rice GI, Kasher PR, Forte GMA, Mannion NM, Greenwood SM, Szynkiewicz M, Dickerson JE, Bhaskar SS, Zampini M, Briggs TA et al (2012) Mutations in ADAR1 cause Aicardi-Goutières syndrome associated with a type I interferon signature. Nat Genet 44:1243–1248
Pichlmair A, Schulz O, Tan CP, Näslund TI, Liljeström P, Weber F, Reis e Sousa C (2006) RIG-I-mediated antiviral responses to single-stranded RNA bearing 5’-phosphates. Science 314:997–1001
Goubau D, Schlee M, Deddouche S, Pruijssers AJ, Zillinger T, Goldeck M, Schuberth C, Van der Veen AG, Fujimura T, Rehwinkel J et al (2014) Antiviral immunity via RIG-I-mediated recognition of RNA bearing 5’-diphosphates. Nature 514:372–375
del Toro Duany Y, Bin W, Sun H (2015) MDA5—filament, dynamics and disease. Curr Opin Virol 12:20–25
Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ et al (2006) Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441:101–105
Hornung V, Ellegast J, Kim S, Brzózka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann K-K, Schlee M et al (2006) 5’-Triphosphate RNA is the ligand for RIG-I. Science 314:994–997
Schlee M, Roth A, Hornung V, Hagmann CA, Wimmenauer V, Barchet W, Coch C, Janke M, Mihailovic A, Wardle G et al (2009) Recognition of 5’ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity 31:25–34
Schmidt A, Schwerd T, Hamm W, Hellmuth JC, Cui S, Wenzel M, Hoffmann FS, Michallet M-C, Besch R, Hopfner K-P et al (2009) 5’-triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. Proc Natl Acad Sci U S A 106:12067–12072
Peisley A, Jo MH, Lin C, Wu B, Orme-Johnson M, Walz T, Hohng S, Hur S (2012) Kinetic mechanism for viral dsRNA length discrimination by MDA5 filaments. Proc Natl Acad Sci U S A 109:E3340–9
Wu B, Peisley A, Richards C, Yao H, Zeng X, Lin C, Chu F, Walz T, Hur S (2013) Structural basis for dsRNA recognition, filament formation, and antiviral signal activation by MDA5. Cell 152:276–289
Rice GI, del Toro Duany Y, Jenkinson EM, Forte GMA, Anderson BH, Giada A, Brigitte B-M, Baildam EM, Roberta B, Beresford MW et al (2014) Gain-of-function mutations in IFIH1 cause a spectrum of human disease phenotypes associated with upregulated type I interferon signaling. Nat Genet 46:503–509
Patterson JB, Samuel CE (1995) Expression and regulation by interferon of a double-stranded-RNA-specific adenosine deaminase from human cells: evidence for two forms of the deaminase. Mol Cell Biol 15:5376–5388
Bazak L, Haviv A, Barak M, Jacob-Hirsch J, Deng P, Zhang R, Isaacs FJ, Rechavi G, Li JB, Eisenberg E et al (2013) A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes. Genome Res 24:365–376
Higuchi M, Miyoko H, Single FN, Martin K, Bernd S, Rolf S, Seeburg PH (1993) RNA editing of AMPA receptor subunit GluR-B: A base-paired intron-exon structure determines position and efficiency. Cell 75:1361–1370
Higuchi M, Maas S, Single FN, Hartner J, Rozov A, Burnashev N, Feldmeyer D, Sprengel R, Seeburg PH (2000) Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature 406:78–81
Behm M, Öhman M (2016) RNA editing: a contributor to neuronal dynamics in the mammalian brain. Trends Genet 32(3):165–75
Rueter SM, Dawson TR, Emeson RB (1999) Regulation of alternative splicing by RNA editing. Nature 399:75–80
Ekdahl Y, Farahani HS, Behm M, Lagergren J, Öhman M (2012) A-to-I editing of microRNAs in the mammalian brain increases during development. Genome Res 22:1477–1487
Vesely C, Tauber S, Sedlazeck FJ, von Haeseler A, Jantsch MF (2012) Adenosine deaminases that act on RNA induce reproducible changes in abundance and sequence of embryonic miRNAs. Genome Res 22:1468–1476
Yang W, Chendrimada TP, Wang Q, Higuchi M, Seeburg PH, Shiekhattar R, Nishikura K (2006) Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat Struct Mol Biol 13:13–21
Whipple JM, Youssef OA, Aruscavage PJ, Nix DA, Hong C, Johnson WE, Bass BL (2015) Genome-wide profiling of the C. elegans dsRNAome. RNA 21:786–800
Osenberg S, Dominissini D, Rechavi G, Eisenberg E (2009) Widespread cleavage of A-to-I hyperediting substrates. RNA 15:1632–1639
Porath HT, Carmi S, Levanon EY (2014) A genome-wide map of hyper-edited RNA reveals numerous new sites. Nat Commun 5:4726
Hundley HA, Bass BL (2010) ADAR editing in double-stranded UTRs and other noncoding RNA sequences. Trends Biochem Sci 35:377–383
Pfaller CK, Li Z, George CX, Samuel CE (2011) Protein kinase PKR and RNA adenosine deaminase ADAR1: new roles for old players as modulators of the interferon response. Curr Opin Immunol 23:573–582
Hartner JC, Schmittwolf C, Kispert A, Müller AM, Higuchi M, Seeburg PH (2004) Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J Biol Chem 279:4894–4902
Wang Q, Miyakoda M, Yang W, Khillan J, Stachura DL, Weiss MJ, Nishikura K (2004) Stress-induced apoptosis associated with null mutation of ADAR1 RNA editing deaminase gene. J Biol Chem 279:4952–4961
Hartner JC, Walkley CR, Lu J, Orkin SH (2009) ADAR1 is essential for the maintenance of hematopoiesis and suppression of interferon signaling. Nat Immunol 10:109–115
Mannion NM, Greenwood SM, Robert Y, Sarah C, James B, David R, Christoffer N, Cornelia V, Ponting CP, McLaughlin PJ et al (2014) The RNA-editing enzyme ADAR1 controls innate immune responses to RNA. Cell Rep 9:1482–1494
Yu S, Sharma R, Nie D, Jiao H, Im H-J, Lai Y, Zhao Z, Zhu K, Fan J, Chen D et al (2013) ADAR1 ablation decreases bone mass by impairing osteoblast function in mice. Gene 513:101–110
Wang G, Wang H, Singh S, Zhou P, Yang S, Wang Y, Zhu Z, Zhang J, Chen A, Billiar T et al (2015) ADAR1 prevents liver injury from inflammation and suppresses interferon production in hepatocytes. Am J Pathol 185:3224–3237
Jiang Q, Crews LA, Barrett CL, Chun H-J, Court AC, Isquith JM, Zipeto MA, Goff DJ, Minden M, Sadarangani A et al (2013) ADAR1 promotes malignant progenitor reprogramming in chronic myeloid leukemia. Proc Natl Acad Sci U S A 110:1041–1046
Pestal K, Kathleen P, Funk CC, Snyder JM, Price ND, Treuting PM, Stetson DB (2015) Isoforms of RNA-editing enzyme ADAR1 independently control nucleic acid sensor MDA5-driven autoimmunity and multi-organ development. Immunity 43:933–944
Ota H, Sakurai M, Gupta R, Valente L, Wulff B-E, Ariyoshi K, Iizasa H, Davuluri RV, Nishikura K (2013) ADAR1 forms a complex with Dicer to promote microRNA processing and RNA-induced gene silencing. Cell 153:575–589
Feng S, Shu F, Heng L, Jing Z, Konstantin P, Ky L, Schwartz TU, Peter D (2011) Alternate rRNA secondary structures as regulators of translation. Nat Struct Mol Biol 18:169–176
Liddicoat BJ, Piskol R, Chalk AM, Ramaswami G, Higuchi M, Hartner JC, Li JB, Seeburg PH, Walkley CR (2015) RNA editing by ADAR1 prevents MDA5 sensing of endogenous dsRNA as nonself. Science 349:1115–1120
Volkman HE, Stetson DB (2014) The enemy within: endogenous retroelements and autoimmune disease. Nat Immunol 15:415–422
Peng Z, Zhiyu P, Yanbing C, Tan BC-M, Lin K, Zhijian T, Yuankun Z, Wenwei Z, Yu L, Xueda H et al (2012) Comprehensive analysis of RNA-Seq data reveals extensive RNA editing in a human transcriptome. Nat Biotechnol 30:253–260
George CX, Gokul R, Li JB, Samuel CE (2016) Editing of cellular self RNAs by adenosine deaminase ADAR1 suppresses innate immune stress responses. J Biol Chem 291(12):6158–68
Ward SV, George CX, Welch MJ, Liou L-Y, Hahm B, Lewicki H, de la Torre JC, Samuel CE, Oldstone MB (2011) RNA editing enzyme adenosine deaminase is a restriction factor for controlling measles virus replication that also is required for embryogenesis. Proc Natl Acad Sci U S A 108:331–336
Acknowledgements
Work in the laboratory was/is supported by The Leukaemia Foundation (CRW); NHMRC Project Grant (CRW; APP1021216 and APP1102006), NHMRC Career Development Award (CRW; APP559016); in part by the Victorian State Government’s OIS Program (to St. Vincent’s Institute); CRW was the Leukaemia Foundation Phillip Desbrow Senior Research Fellow.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Author contribution
J.E.H-F. and C.R.W. analysed and interpreted data, provided intellectual input and wrote the manuscript.
Conflict of interest
The funders of our work had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript. All authors declare no conflicts of interest.
Rights and permissions
About this article
Cite this article
Heraud-Farlow, J.E., Walkley, C.R. The role of RNA editing by ADAR1 in prevention of innate immune sensing of self-RNA. J Mol Med 94, 1095–1102 (2016). https://doi.org/10.1007/s00109-016-1416-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00109-016-1416-1