Abstract
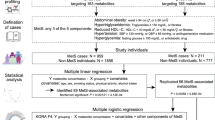
Chronic cortisol excess induces several alterations on protein, lipid and carbohydrate metabolism resembling those found in the metabolic syndrome. However, patients exposed to prolonged high levels of cortisol in Cushing syndrome (CS) present exceeding cardiometabolic alterations not reflected by conventional biomarkers. Using 3 ultra-high-performance liquid chromatography-tandem mass spectrometry (UHPLC-MS) platforms, we aimed to characterise the serum metabolome of 25 patients with active endogenous CS and 25 control subjects matched by propensity score (sex, BMI, diabetes mellitus type 2 (T2D), high blood pressure (HBP) and dyslipidaemia) to search for potential disease-specific biomarkers and pathways associated to the clinical comorbidities. A total of 93 metabolites were significantly altered in patients with CS. Increased levels of sulfur amino acids (AA), triacylglycerols, glycerophospholipids, ceramides and cholesteryl esters were observed. Contrarily, concentrations of essential and non-essential AA, polyunsaturated fatty acids, conjugated bile acids and second messenger glycerolipids were decreased. Twenty-four-hour urinary free cortisol (24h-UFC) independently determined the concentration of 21 lipids and 4 AA. A metabolic signature composed by 10 AA and 10 lipid metabolites presented an AUC-ROC of 95% for the classification of CS patients. Through differential network analysis, 152 aberrant associations between metabolites involved in the Lands cycle and Kennedy pathway were identified. Our data indicates that chronic hypercortisolemia confers a unique lipidomic signature and several alterations in numerous AA even when compared to patients with similar metabolic comorbidities providing novel insights of the increased cardiometabolic burden of CS.
Key messages
• Cortisol excess induces metabolic alterations beyond conventional biomarkers.
• The hypercortisolism extent determines the concentration of 21 lipids and 5 aa.
• Cortisol excess confers a unique metabolic signature of 20 metabolites.
• Kennedy and Lands cycle are profoundly disturbed by cortisol excess.





Similar content being viewed by others
Data availability
All data, materials and software application or custom code support their published claims and comply with field standards. All metabolomic data generated and analysed during this study is included in the present manuscript and as supplementary spreadsheets material in the initial submission (Additional File 2). The clinical dataset of the participants included in the study is available from the corresponding author upon reasonable request. Codes employed in R (programming language) for the present study are available from the corresponding author upon reasonable request.
References
Lacroix A, Feelders RA, Stratakis CA, Nieman LK (2015) Cushing’s syndrome. Lancet 386:913–927
Ntali G, Asimakopoulou A, Siamatras T, Komninos J, Vassiliadi D, Tzanela M, Tsagarakis S, Grossman AB, Wass JAH, Karavitaki N (2013) Mortality in Cushing’s syndrome: systematic analysis of a large series with prolonged follow-up. Eur J Endocrinol 169:715–723
Chanson P, Salenave S (2010) Metabolic syndrome in Cushing’s syndrome. Neuroendocrinology 92:96–101
Pivonello R, Isidori AM, De Martino MC et al (2016) Complications of Cushing’s syndrome: state of the art. Lancet Diabetes Endocrinol 4:611–629
Di Dalmazi G, Quinkler M, Deutschbein T et al (2017) Cortisol-related metabolic alterations assessed by mass spectrometry assay in patients with Cushing’s syndrome. Eur J Endocrinol 177:227–237
Faggiano A, Pivonello R, Melis D, Alfieri R, Filippella M, Spagnuolo G, Salvatore F, Lombardi G, Colao A (2002) Evaluation of circulating levels and renal clearance of natural amino acids in patients with Cushing’s disease. J Endocrinol Investig 25:142–151
Lu Y, Zhang Z, Xiong X, Wang X, Li J, Shi G, Yang J, Zhang X, Zhang H, Hong J, Xia X, Ning G, Li X (2012) Glucocorticoids promote hepatic cholestasis in mice by inhibiting the transcriptional activity of the farnesoid X receptor. Gastroenterology 143:1630–1640
Faggiano A, Melis D, Alfieri R, de Martino MC, Filippella M, Milone F, Lombardi G, Colao A, Pivonello R (2005) Sulfur amino acids in Cushing’s disease: insight in homocysteine and taurine levels in patients with active and cured disease. J Clin Endocrinol Metab 90:6616–6622
Zamboni N, Saghatelian A, Patti GJ (2015) Defining the metabolome: size, flux, and regulation. Mol Cell 58:699–706
Nieman LK, Biller BMK, Findling JW, Newell-Price J, Savage MO, Stewart PM, Montori VM (2008) The diagnosis of Cushing’s syndrome: an endocrine society clinical practice guideline. J Clin Endocrinol Metab 93:1526–1540
Centro de investigacion biomedica en red I de salud C (2015) Understanding obesity (Ob), metabolic syndrome (MetS), type 2 diabetes (T2DM) and fatty liver disease (FL): a multidisciplinary approach. In: Proy. Integr. Excel. – Convoc. AES2014. https://www.ciberisciii.es/media/1002074/ciberehd_understandingob.pdf. Accessed 14 Feb 2020
Tokarz J, Haid M, Cecil A, Prehn C, Artati A, Möller G, Adamski J (2017) Endocrinology meets metabolomics: achievements, pitfalls, and challenges. Trends Endocrinol Metab 28:705–721
Austin PC (2011) An introduction to propensity score methods for reducing the effects of confounding in observational studies. Multivar Behav Res 46:399–424
Aranda G, Fernandez-Ruiz R, Palomo M, Romo M, Mora M, Halperin I, Casals G, Enseñat J, Vidal O, Diaz-Ricart M, Hanzu FA (2018) Translational evidence of prothrombotic and inflammatory endothelial damage in Cushing syndrome after remission. Clin Endocrinol 88:415–424
Aranda G, Careaga M, Hanzu FA, Patrascioiu I, Ríos P, Mora M, Morales-Romero B, Jiménez W, Halperin I, Casals G (2016) Accuracy of immunoassay and mass spectrometry urinary free cortisol in the diagnosis of Cushing’s syndrome. Pituitary 19:496–502
Wagner C, Veyrie N, Martín-Duce A et al (2010) Liquid chromatography−mass spectrometry-based parallel metabolic profiling of human and mouse model serum reveals putative biomarkers associated with the progression of nonalcoholic fatty liver disease. J Proteome Res 9:4501–4512
Fernández-Escalante C, Clement K, Andrade RJ et al (2012) Obesity-dependent metabolic signatures associated with nonalcoholic fatty liver disease progression. J Proteome Res 11:2521–2532
Van Meer G, Voelker DR, Feigenson GW (2008) Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol 9:112–124
Mayo R, Alonso C, Mato JM et al (2015) Enhancing metabolomics research through data mining. J Proteome 127:275–288
Van Der Kloet FM, Bobeldijk I, Verheij ER, Jellema RH (2009) Analytical error reduction using single point calibration for accurate and precise metabolomic phenotyping. J Proteome Res 8:5132–5141
Wiedermann W, Hagmann M, von Eye A (2015) Significance tests to determine the direction of effects in linear regression models. Br J Math Stat Psychol 68:116–141
Hu T, Zhang W, Fan Z et al (2016) Metabolomics differential correlation network analysis of osteoarthritis. Pac Symp Biocomput:120–131. https://doi.org/10.1142/9789814749411_0012
Li Z, Zhang Y, Hu T, Likhodii S, Sun G, Zhai G, Fan Z, Xuan C, Zhang W (2018) Differential metabolomics analysis allows characterization of diversity of metabolite networks between males and females. PLoS One 13:1–10
Ferraù F, Korbonits M (2015) Metabolic comorbidities in Cushing’s syndrome. Eur J Endocrinol 173:M133–M157
Ong SLH, Whitworth JA (2011) How do glucocorticoids cause hypertension: role of nitric oxide deficiency, oxidative stress, and eicosanoids. Endocrinol Metab Clin N Am 40:393–407
Naray-Fejes-Tóth A, Rosenkranz B, Frölich JC, Fejes-Tóth G (1988) Glucocorticoid effect on arachidonic acid metabolism in vivo. J Steroid Biochem 30:155–159
Serhan CN, Yacoubian S, Yang R (2008) Anti-inflammatory and proresolving lipid mediators. Annu Rev Pathol Mech Dis 3:279–312
Hasan AU, Ohmori K, Hashimoto T, Kamitori K, Yamaguchi F, Rahman A, Tokuda M, Kobori H (2018) PPARγ activation mitigates glucocorticoid receptor-induced excessive lipolysis in adipocytes via homeostatic crosstalk. J Cell Biochem 119:4627–4635
Arnaldi G, Scandali VM, Trementino L, Cardinaletti M, Appolloni G, Boscaro M (2010) Pathophysiology of dyslipidemia in Cushing’s syndrome. Neuroendocrinology 92:86–90
Dolinsky VW, Douglas DN, Lehner R, Vance DE (2004) Regulation of the enzymes of hepatic microsomal triacylglycerol lipolysis and re-esterification by the glucocorticoid dexamethasone. Biochem J 378:967–974
Vance JE, Tasseva G (2013) Formation and function of phosphatidylserine and phosphatidylethanolamine in mammalian cells. Biochim Biophys Acta Mol Cell Biol Lipids 1831:543–554
Liu X, Zheng P, Zhao X, Zhang Y, Hu C, Li J, Zhao J, Zhou J, Xie P, Xu G (2015) Discovery and validation of plasma biomarkers for major depressive disorder classification based on liquid chromatography-mass spectrometry. J Proteome Res 14:2322–2330
Meikle PJ, Wong G, Barlow CK, Weir JM, Greeve MA, MacIntosh GL, Almasy L, Comuzzie AG, Mahaney MC, Kowalczyk A, Haviv I, Grantham N, Magliano DJ, Jowett JBM, Zimmet P, Curran JE, Blangero J, Shaw J (2013) Plasma lipid profiling shows similar associations with prediabetes and type 2 diabetes. PLoS One 8:e74341
Stegemann C, Pechlaner R, Willeit P, Langley SR, Mangino M, Mayr U, Menni C, Moayyeri A, Santer P, Rungger G, Spector TD, Willeit J, Kiechl S, Mayr M (2014) Lipidomics profiling and risk of cardiovascular disease in the prospective population-based bruneck study. Circulation 129:1821–1831
Law S-H, Chan M-L, Marathe GK, Parveen F, Chen CH, Ke LY (2019) An updated review of lysophosphatidylcholine metabolism in human diseases. Int J Mol Sci 20:1149
Gibellini F, Smith TK (2010) The Kennedy pathway-de novo synthesis of phosphatidylethanolamine and phosphatidylcholine. IUBMB Life 62:414–428
Boulgaropoulos B, Amenitsch H, Laggner P, Pabst G (2010) Implication of sphingomyelin/ceramide molar ratio on the biological activity of sphingomyelinase. Biophys J 99:499–506
Holland WL, Brozinick JT, Wang LP, Hawkins ED, Sargent KM, Liu Y, Narra K, Hoehn KL, Knotts TA, Siesky A, Nelson DH, Karathanasis SK, Fontenot GK, Birnbaum MJ, Summers SA (2007) Inhibition of ceramide synthesis ameliorates glucocorticoid-, saturated-fat-, and obesity-induced insulin resistance. Cell Metab 5:167–179
Xia JY, Morley TS, Scherer PE (2014) The adipokine/ceramide axis: key aspects of insulin sensitization. Biochimie 96:130–139
Kurz J, Parnham MJ, Geisslinger G, Schiffmann S (2019) Ceramides as novel disease biomarkers. Trends Mol Med 25:20–32
Chaurasia B, Summers SA (2015) Ceramides - lipotoxic inducers of metabolic disorders. Trends Endocrinol Metab 26:538–550
Brunkhorst-Kanaan N, Klatt-Schreiner K, Hackel J, Schröter K, Trautmann S, Hahnefeld L, Wicker S, Reif A, Thomas D, Geisslinger G, Kittel-Schneider S, Tegeder I (2019) Targeted lipidomics reveal derangement of ceramides in major depression and bipolar disorder. Metabolism 95:65–76
Laaksonen R, Ekroos K, Sysi-Aho M, Hilvo M, Vihervaara T, Kauhanen D, Suoniemi M, Hurme R, März W, Scharnagl H, Stojakovic T, Vlachopoulou E, Lokki ML, Nieminen MS, Klingenberg R, Matter CM, Hornemann T, Jüni P, Rodondi N, Räber L, Windecker S, Gencer B, Pedersen ER, Tell GS, Nygård O, Mach F, Sinisalo J, Lüscher TF (2016) Plasma ceramides predict cardiovascular death in patients with stable coronary artery disease and acute coronary syndromes beyond LDL-cholesterol. Eur Heart J 37:1967–1976
Hilvo M, Meikle PJ, Pedersen ER, Tell GS, Dhar I, Brenner H, Schöttker B, Lääperi M, Kauhanen D, Koistinen KM, Jylhä A, Huynh K, Mellett NA, Tonkin AM, Sullivan DR, Simes J, Nestel P, Koenig W, Rothenbacher D, Nygård O, Laaksonen R (2020) Development and validation of a ceramide- and phospholipid-based cardiovascular risk estimation score for coronary artery disease patients. Eur Heart J 41:371–380
Eloranta JJ, Jung D, Kullak-Ublick GA (2006) The human Na+-taurocholate cotransporting polypeptide gene is activated by glucocorticoid receptor and peroxisome proliferator-activated receptor-γ coactivator-1α, and suppressed by bile acids via a small heterodimer partner-dependent mechanism. Mol Endocrinol 20:65–79
Haeusler RA, Astiarraga B, Camastra S, Accili D, Ferrannini E (2013) Human insulin resistance is associated with increased plasma levels of 12a-hydroxylated bile acids. Diabetes 62:4184–4191
Ahmad TR, Haeusler RA (2019) Bile acids in glucose metabolism and insulin signalling — mechanisms and research needs. Nat Rev Endocrinol 15:701–712
García-Eguren G, Giró O, Romero MDM, Grasa M, Hanzu FA (2019) Chronic hypercortisolism causes more persistent visceral adiposity than HFD-induced obesity. J Endocrinol 242:65–77
Lee IT, Atuahene A, Egritag HE, Wang L, Donovan M, Buettner C, Geer EB (2019) Active Cushing disease is characterized by increased adipose tissue macrophage presence. J Clin Endocrinol Metab 104:2453–2461
Rui WS, Adam J, Brandmaier S et al (2016) Metformin effect on nontargeted metabolite profiles in patients with type 2 diabetes and in multiple murine tissues. Diabetes 65:3776–3785
Würtz P, Wang Q, Soininen P, Kangas AJ, Fatemifar G, Tynkkynen T, Tiainen M, Perola M, Tillin T, Hughes AD, Mäntyselkä P, Kähönen M, Lehtimäki T, Sattar N, Hingorani AD, Casas JP, Salomaa V, Kivimäki M, Järvelin MR, Davey Smith G, Vanhala M, Lawlor DA, Raitakari OT, Chaturvedi N, Kettunen J, Ala-Korpela M (2016) Metabolomic profiling of statin use and genetic inhibition of HMG-CoA reductase. J Am Coll Cardiol 67:1200–1210
Acknowledgements
Arturo Vega-Beyhart sincerely acknowledges the Mexican National Council of Science and Technology (CONACYT) for the scholarship granted to pursue his doctorate at the University of Barcelona.
Funding
This work was supported by 2 grants of the Carlos III Health Institute of MINECO, Spain (FIS PI1500859 and PIE14/00031).
Author information
Authors and Affiliations
Contributions
Conceptualization: Felicia A Hanzu, Adriana Pané, Ramon Gomis, Gemma Rojo, Arturo Vega-Beyhart
Data: Felicia A Hanzu, Arturo Vega-Beyhart, Adriana Pané, Guillermo García-Eguren, Oriol Giró, Laura Boswell, Gloria Aranda, Vanesa Flores, Francesc Carmona, Joaquim Enseñat, Oscar Vidal, Mireia Mora, Irene Halperin, Gemma Rojo
Formal analysis: Felicia A Hanzu, Arturo Vega-Beyhart, Marta Iruarrizaga, Ting Hu, Guillermo García-Eguren, Oriol Giró, Gregori Casals
Funding acquisition: Felicia A Hanzu, Gemma Rojo, Ramon Gomis
Investigation: All authors participated
Supervision: Felicia A Hanzu
Writing, review and editing: All authors participated
Corresponding author
Ethics declarations
Ethics approval
The present study followed international and national regulations including the Declaration of Helsinki and national notes for guidance on Good Clinical Practice CPMP/ICH/135/95. The Institutional Research and Ethics Committee at Hospital Clínic de Barcelona approved the present study (Comité de Ética de la Investigación con medicamentos del Hospital Clínic de Barcelona). The Institutional Research and Ethics Committee at Hospital Regional Universitario de Málaga approved the present study (Comité de Ética de la Investigación Provincial de Málaga).
Consent to participate
Written informed consent was obtained from all participants prior to study inclusion and are available under considerable request.
Consent for publication
Not applicable. No personal nor any clinical details of participants are presented.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Vega-Beyhart, A., Iruarrizaga, M., Pané, A. et al. Endogenous cortisol excess confers a unique lipid signature and metabolic network. J Mol Med 99, 1085–1099 (2021). https://doi.org/10.1007/s00109-021-02076-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00109-021-02076-0